| Citation: | Yongbin Chang, Gang Song, Dezhi Zhang, Chenxi Jia, Ping Fan, Yan Hao, Yanzhu Ji, Fumin Lei. 2022: Distribution pattern and driving factors of genetic diversity of passerine birds in the Mountains of Southwest China. Avian Research, 13(1): 100043. DOI: 10.1016/j.avrs.2022.100043 |
Genetic diversity is one of the three dimensions of biodiversity and fundamental to various life forms on the Earth. Understanding the distribution pattern of genetic diversity and its driving forces has been an important topic in ecology, biogeography and conservation biology since the last decade. We investigated the genetic diversity pattern of passerine birds in the Mountains of Southwest China, a global biodiversity hotspot with the highest species richness of birds in the entire Eurasia, and explored the influencing forces of environmental variables on genetic diversity. We compiled 1189 Cytochrome b sequences of 27 passerine species from 152 geographic sites, covering the range of Mountains of Southwest China and its adjoining areas. We generated genetic diversity distribution maps using a grid-cell method based on nucleotide diversity and haplotype diversity indices. We further analyzed the variation pattern of the two indices along latitudinal, longitudinal, and elevational gradients. The correlations between the two indices and environmental variables were also evaluated. The nucleotide diversity hotspots were mostly located in the southern Hengduan Mountains, while for haplotype diversity, three hotspots were detected: the southeast edge of the Qinghai-Tibetan Plateau, the southern Hengduan Mountains and the Qinling Mountains. There was no monotonic increasing or decreasing pattern in nucleotide diversity or haplotype diversity along latitudinal, longitudinal or elevational gradients except for altitudinal range. Correlation and model selection analyses detected multiple environmental variables in driving genetic diversity patterns, including temperature, precipitation, vegetation, human influence, longitude and altitude range. Similar to the pattern of species richness, the nucleotide diversity pattern of passerine birds in the Mountains of Southwest China presents a decreasing trend from southwest to northeast, while the haplotype diversity pattern is more likely decreased from west to east. Our results indicate that the distribution pattern of genetic diversity may be derived from the complex topography and diverse microclimates in the Mountains of Southwest China.
As the fundamental component of biodiversity, genetic diversity (GD) plays a vital role in forming the variety of natural organisms on the Earth (Heywood and Watson, 1995). GD is defined as any measurement that quantifies the magnitude of genetic variability within a population (Fischer, 1930). It is a quantitative index of the evolutionary potential of a population that aids population's adaptability in response to spatial and temporal variation (Wei et al., 2017). Nucleotide diversity (π) and haplotype diversity (Hd) are two indices profiling the genetic polymorphism of populations. Nucleotide diversity is defined as the number of different nucleotide sites between two randomly chosen alleles, while Hd represents the probability that two randomly sampled alleles are different (Nei and Li, 1979; Nei, 1987). Nucleotide diversity represents the cumulative degree of genetic variation during evolution, while Hd reflects the particular combination of haplotypes, which is more relevant to population dynamics at short time-scales (Pauls et al., 2013; Leitwein et al., 2020). Many studies have demonstrated the importance of GD in ecology and evolution, but most of them have mainly focused on a handful of species or populations (Xu et al., 2018; Leigh et al., 2021).
In recent decades, owning to public databases of a large number of DNA sequences, it become feasible to analyze the global GD pattern based on multiple taxa. After a pioneer study on the global GD patterns of terrestrial vertebrates (Miraldo et al., 2016), many studies on GD patterns at the global scale have emerged for mammals and amphibians (Gratton et al., 2017), fishes (Manel et al., 2020), insects and plants (Millette et al., 2019). These studies applied gene markers of either neutral genes, such as Cytochrome b (Cyt b) and Cytochrome Oxidase I (COI), or functional genes, such as major histocompatibility complex genes (MHC II) (Li et al., 2021). However, most of these studies focused on macro-scaled ranges, e.g., at a country or continent level (Miraldo et al., 2016; Millette et al., 2019; Hu et al., 2021; Schmidt and Garroway, 2021). Research efforts at smaller regional scales and/or at species divergence centres are rather scarce (Wang et al., 2013; Deng et al., 2019; Yu et al., 2019).
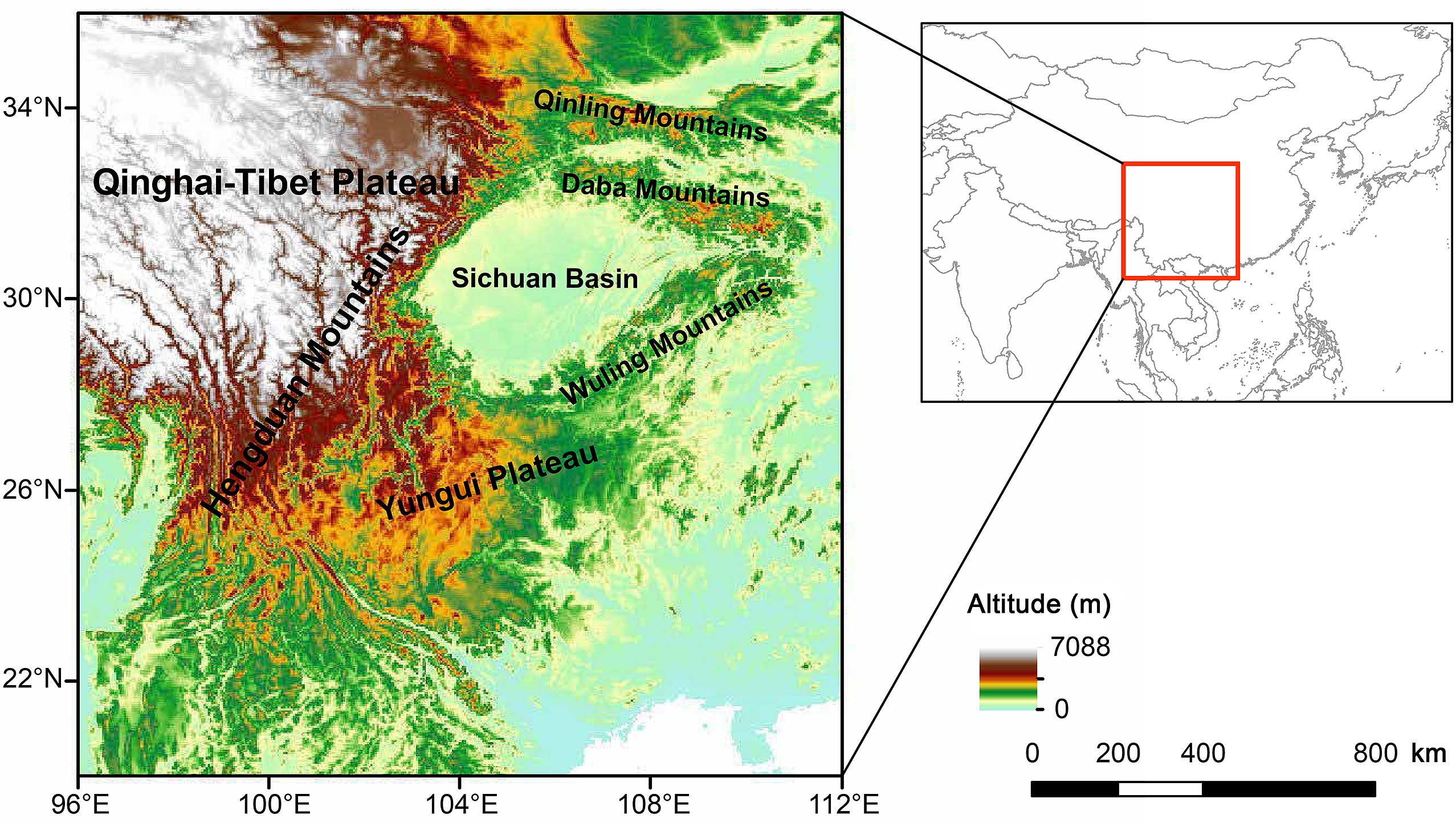
As one of the 35 global biodiversity hotspots, the Mountains of Southwest China (MSC) harbor extraordinary fauna and flora within a comparatively narrow land area (Myers et al., 2000; Mittermeier et al., 2011). The MSC is characterized by high landscape complexity and vertical variation. There are 939 bird species in this area, representing 64% of all bird species in China, ranking the highest avian species richness region in China (Wu et al., 2017). It provided multiple micro-refugia for many species during the Last Glacial Maximum (LGM) despite of many glaciers distributed in this area (He and Jiang, 2014; Lei et al., 2015). For example, a previous study revealed that speciation events in Leaf Warblers (Phylloscopus spp.) were attributed to dispersal into the Himalayas followed by vicariance between the Himalayas and China/Southeast Asia before the Pleistocene (Johansson et al., 2007). Some studies on avian phylogeography related to this region found multiple lineages within such a narrow area (Song et al., 2009; Liu et al., 2012). Recent studies also show that this region is the centre of speciation for several avian assemblages, e.g., Phasianidae, Paridae, and Leiothrichidae (Fjeldså, 2013; Cai et al., 2018, 2020; Johansson et al., 2018). The important role of topographic features of MSC in shaping the lineage diversification of many avian groups has been illustrated (Reddy and Moyle, 2011; Lei et al., 2015). However, the general GD distribution pattern and its driving factors are still unknown regard to this region.
Therefore, in this study, we performed a meta-analysis with abundant of sequences from multiple passerine species (mostly forest- and bush-dwelling birds) in the MSC region. We aimed to investigate: 1) whether there is a geographic gradient of GD along latitudinal, longitudinal and elevational gradients; 2) whether GD shows a similar pattern to species diversity in this region; and 3) which environmental variables drive the GD distribution patterns.
The MSC includes several mountain ranges: the Hengduan Mountains, Qinling Mountains, Daba Mountains, Yungui Plateau, Wuling Mountains and the southeast edge of the Qinghai-Tibetan Plateau (QTP) (Wu et al., 2013, 2017). This region is one of the world's 35 biodiversity hotspots and has the highest biodiversity in China (Mittermeier et al., 2011). Our study area focused on mountainous areas spanning 96° E−112° E and 20° N–36° N, which includes the major ranges of the MSC and adjacent areas (Fig. 1).
The MSC is prominent in its avian species richness, particularly the species richness and endemicity of passerine birds (Lei et al., 2003; Wu et al., 2017). Substantial phylogeographical studies of dozens of passerine birds in the MSC have been conducted (Appendix Table S1), which provide plenty of sequences for the study of population genetics. Therefore, we focused on passerine species in this study.
We selected the mitochondrial Cyt b fragment as the genetic marker for GD calculation in this study, as Cyt b is one of the most popular gene markers in phylogeographical studies and a large number of Cyt b sequences have been deposited in public databases. In total, we compiled 1189 sequences from 27 passerine species across 152 sampling sites (Appendix Table S1). Among them, 1137 sequences of 24 species were obtained from previous studies (Appendix Table S1), 51 sequences of 4 species were amplified and sequenced in this study following the experimental protocol in a previous study (Zhao et al., 2019). The newly generated sequences were deposited in GenBank (Accession No. OL906330–OL906380).
To investigate the relationship between GD and environmental variables, data on 19 bioclimatic variables and altitude were downloaded from the WorldClim database (http://www.worldclim.org/) (Hijmans et al., 2005), normalized difference vegetation index (NDVI) and enhanced vegetation index (EVI) data were downloaded from the Geospatial Data Cloud site of the Computer Network Information Centre, Chinese Academy of Sciences (http://www.gscloud.cn). In order to estimate whether past climate oscillations had dramatic effect on the genetic diversity, we also downloaded the distribution map of glacier sheets during the LGM from the Collaborative Research Centre 806 Database (https://crc806db.uni-koeln.de/layer/show/6). The Human Influence Index (HII) data were downloaded from the Socioeconomic Data and Applications Centre (http://sedac.ciesin.columbia.edu/wildareas/). A total of 26 environmental variables were included in this study (Appendix Table S2).
All Cyt b sequences were aligned in Mega 6.0 (Tamura et al., 2013). Each sequence was annotated with coordinates according to its original sampling information. For each population of every species from the sampling site, we calculated two parameters (π and Hd) in DNASP 5.10 (Librado and Rozas, 2009). Populations with a sample size of less than three individuals were removed according to the calculation specifications of DNASP 5.10. Finally, we obtained 152 geographic sites with GD values for downstream analyses (Appendix Table S3).
We divided the study area into 1 × 1° grids for a total of 256 grid cells (16 × 16). We calculated the mean value of π and Hd for each grid cell in ArcGIS 10.0 (ESRI, Redlands, CA, USA). For example, if two or more geographic points were located in one cell, we calculated the mean values of π and Hd from these points. After distributing the population GD values into the grid cells, we obtained 60 cells with validated values. To better predict the pattern of GD distribution, we applied the hypothesis of continuous spreading of the population by interpolating data from validated grid cells to adjacent areas. The π and Hd values of each grid cell were interpolated by the Kriging method (Oliver and Webster, 1990) in the ArcGIS spatial analysis toolbox.
We extracted the mean values of 26 environmental variables for each grid cell (Appendix Table S4). To detect the effects of habitat heterogeneity, we calculated the altitude range (altRange, the highest altitude value minus the lowest altitude value) within every grid cell. Environmental variables extraction was performed in ArcGIS 10.0. Considering the correlation between variables, we first performed Pearson's correlation tests (Song et al., 2018) to select the essential environmental variables. When the correlation coefficient between two variables was higher than 0.8, we chose the variable that was more biologically meaningful and more commonly used in biodiversity studies, such as BIO1 (annual mean temperature), BIO7 (annual temperature range), and BIO12 (annual precipitation), in subsequent analyses (Smith et al., 2017; Abebe et al., 2019).
A Spearman's rank correlation analysis was performed between GD values and these environmental variables to detect the most correlated variables. A model selection analysis was also conducted to find the best explanatory variables for GD with a corrected Akaike Information Criterion (AIC) value (Anderson et al., 1998). To check the geographical patterns of GD, we performed locally weighted regression (LOESS) analyses for π and Hd along latitudinal, longitudinal and altitudinal gradients, as well as altitudinal range of each grid cell, which represents the topographical complexity. Ordinary least-squares (OLS) linear regression was also conducted for comparison.
Pearson's and Spearman's correlation analyses were performed in SPSS 18.0 (SPSS, IL, USA). The model selection analysis and Moran's I test (Moran, 1950) for spatial autocorrelation were conducted in SAM 4.0 (Rangel et al., 2010). The OLS linear regression and LOESS were visualized in the ggplot2 package in R studio (Wickham, 2016) with R v. 4.0.2 (R Core Team, 2020).
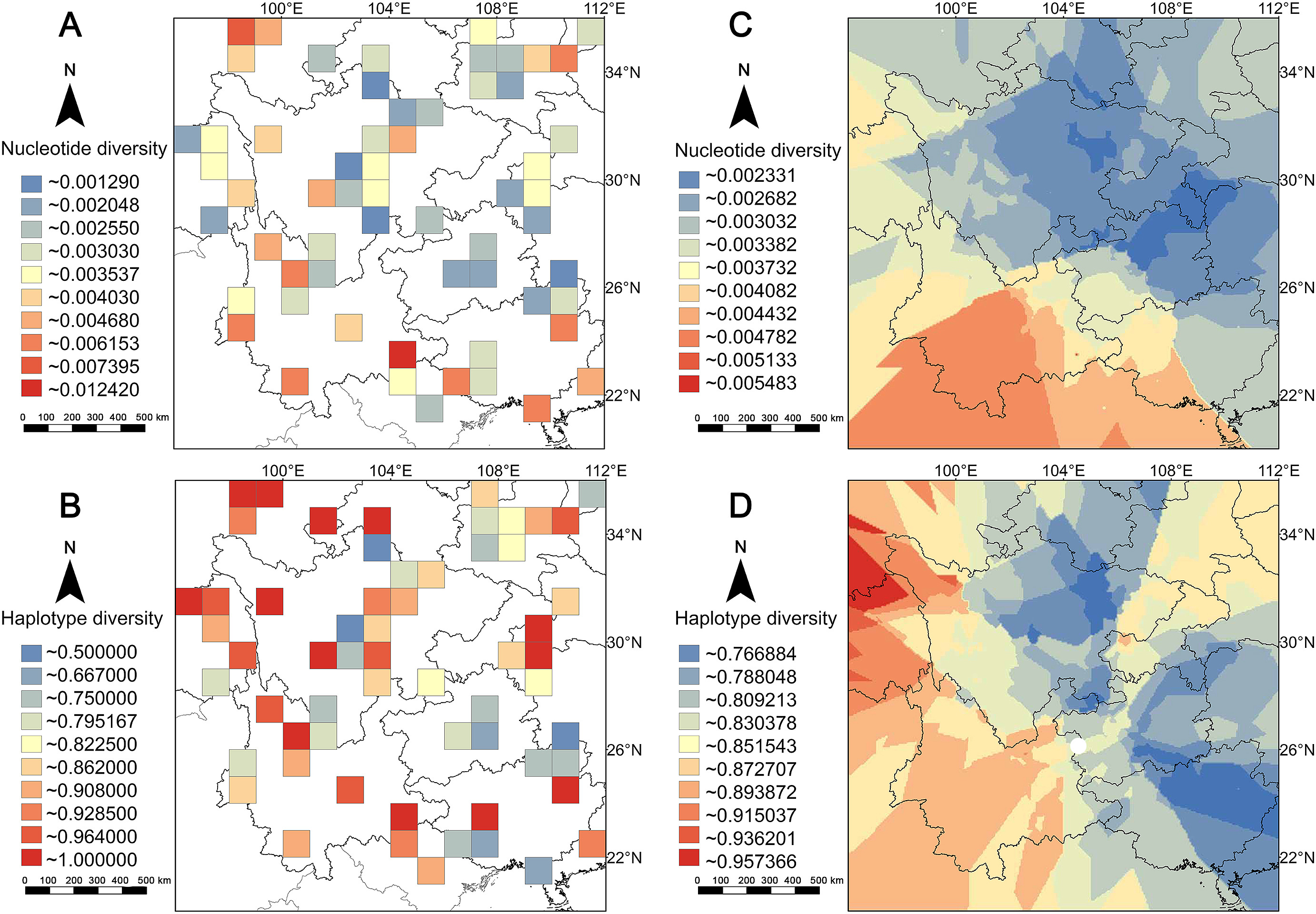
Among the 152 populations in this study, only 15.1% (23/152) of the populations had π values higher than 0.005, while 49.3% (75/152) of the populations had Hd values higher than 0.9. We found 33 populations with Hd values of 1.0, which indicated that every Cyt b sequence from those populations was unique. Among the 60 cells in the dataset, 15% (9/60) had π values higher than 0.005, 46.7% (28/60) Hd values greater than 0.9, and 12 cells had an Hd value of 1.0 (Appendix Table S4).
The interpolation analysis found that the area with the highest π value was located in the southern Hengduan Mountains (Fig. 2C), which was consistent with the results from the cell data analysis (Fig. 2A). Among the nine cells with higher values (π ≥ 0.005), seven cells were located in the South China zoogeographical region (Appendix Fig. S1A), which is characterized by a tropical lowland forest geographical feature. One of the remaining two cells was found in the Qinling Mountains, which is the boundary between the Palearctic and the Oriental realms. The other one was located on the southeast edge of the QTP. Interestingly, we found a similar division of higher (π ≥ 0.005)/lower (π < 0.005) π value cells at the boundary between the Central China/Southwest China and South China zoogeographical regions (Zhang, 1999) (Appendix Fig. S1). Overall, our results showed that the southwest part of the MSC had a higher level of nucleotide diversity.
The Hd pattern was different than the π pattern. The cells with higher values (Hd ≥ 0.9) were broadly distributed on the southeast QTP, Hengduan, Wuling and Qingling Mountains. Unlike patterns of the π values, no clear divisions in Hd values were identified (Fig. 2B; Appendix Fig. S1B). When comparing the patterns of GD with the distribution of LGM glacier, we found that the middle to low values of π in the grid cells were distributed in the glacier-covered areas, while for Hd, both high and low values were recorded in the region under glacier cover (Appendix Fig. S2).
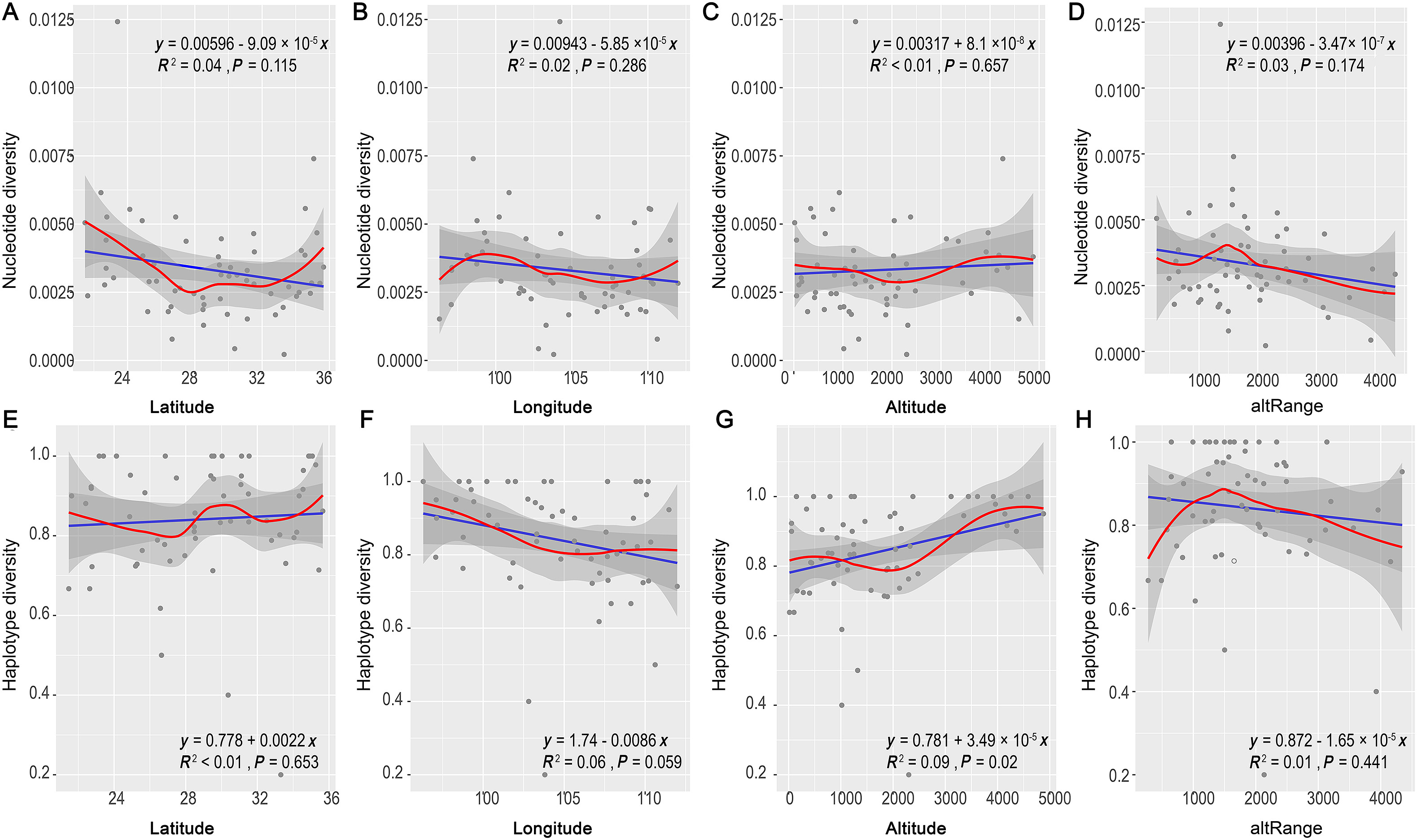
The local weighted regression showed a nearly "W"-shaped pattern of π along the latitudinal gradient. The π value decreased from 20° N to 28° N, reached its lowest point, then increased from 28° N to 29° N, slightly decreased between 29 and 32° N, hit the second bottom near 32° N, and then increased to 36° N (Fig. 3A). The Hd value along the latitudinal gradient showed a similar "W"-shaped pattern, with two bottoms at 28° N and 32° N and a peak at 30° N (Fig. 3E). We observed an "S" variation pattern of π values along the longitudinal gradient, with the peak near 99° E and a bottom near 107° E (Fig. 3B). The Hd value showed a decreasing pattern from 96 to 105° E and then slightly increased from 105° E to 112° E (Fig. 3F). For altitude, the π value showed a "V" pattern with its lowest point near 2000 m. The curve slightly decreased below 1000 m, increased more steeply from 2000 to 4000 m and reached a peak at 4000 m. (Fig. 3C). Hd showed an "M" pattern, first reaching a peak near 500 m, then decreasing from 500 to 2000 m and reaching its lowest point at 2000 m, then distinctly increased from 2000 to 4500 m and followed by a weak decrease to 5000 m. (Fig. 3G). Among all the regression tests, only the linear regression for Hd along the altitudinal gradient showed significance (p = 0.02; Fig. 3G), indicating a positive relationship between Hd and elevation. We observed a consistent unimodal trend both for π and Hd with the highest value at altRange around 1500 m (from 1000 to 2000; Fig. 3D, H).
After removing highly correlated environmental variables (Appendix Table S5), 13 variables remained (Table 1). Spearman's correlation tests showed that the correlations with environmental variables were different for π and Hd. Nucleotide diversity was positively correlated with isothermality (BIO3; r = 0.315, p = 0.016) and precipitation seasonality (BIO15; r = 0.279, p = 0.034). Hd was positively correlated with precipitation seasonality (BIO15; r = 0.271, p = 0.040).
| Spearman rho | π | Hd |
| BIO1 | 0.088 | -0.195 |
| BIO3 | 0.315* | 0.199 |
| BIO4 | -0.196 | -0.057 |
| BIO7 | -0.135 | 0.094 |
| BIO12 | -0.034 | -0.207 |
| BIO15 | 0.279* | 0.217* |
| NDVI | -0.093 | -0.234 |
| EVI | 0.084 | 0.243 |
| HII | 0.027 | -0.143 |
| Altitude | 0.021 | 0.236 |
| altRange | -0.100 | -0.120 |
| Longitude | -0.160 | -0.218 |
| Latitude | -0.111 | 0.098 |
| π, nucleotide diversity, Hd, haplotype diversity. *P < 0.05, **P < 0.01. | ||
Spatial autocorrelation analysis found that the π value was spatially auto-correlated with the cell datasets, and the autocorrelation should be taken into account in the subsequent model selection. The model selection analysis found that the most correlated environmental variables for π were NDVI, HII and longitude. For Hd, only BIO15 and altRange were selected (Table 2).
| Index | Selection variables |
| π | NDVI, HII, longitude |
| Hd | BIO15, altRange |
Genetic diversity plays a fundamental role in the diversification of the world's organisms. Understanding the distribution pattern of global GD and its relationship to biodiversity is an emerging research topic in ecology and biogeography (Leigh et al., 2021). Fortunately, many phylogeographical studies have noted this important dimension of biodiversity in the MSC (Wan et al., 2021). Leveraging a substantial amount of sequence data from birds of the MSC, our results contribute to the understanding of the distribution pattern of GD and the environmental factors driving its evolution. Whether GD has been driven by similar environmental factors underlying species richness is controversial (Lawrence and Fraser, 2020). This question particularly warrants a further research effort in the MSC, where many endemic species and cryptic genetic lineages have been frequently discovered (Wu et al., 2017; Wan et al., 2021).
The distribution patterns of π and Hd are different, possibly due to the intrinsic properties of these two parameters. Nucleotide diversity is more relevant to the cumulative degree of genetic variation along time, while Hd reflects the particular combination of haplotypes, which is more sensitive to population dynamics in a short-time history, such as changes in allele frequency (Pauls et al., 2013; Leitwein et al., 2020). This is in line with the GD patterns in area under the LGM glacier-cover, where we found low values of π but both high and low values of Hd (Appendix Fig. S2). The lower level of π indicates the glacier impacts on historical population (Cheng et al., 2021), while the high Hd values implies rapid post-glacial demographic expansion from those refugia populations.
We have noticed that the higher π value is mainly located in the southern China zoogeographical region, corresponding to two biodiversity hotspots, the MSC and Indo-Burma hotspots (Lei et al., 2015). There are also two high-diversity cells located in the Qinling Mountains and the southeast edge of the QTP. The continuous interpolation pattern shows that the southern Hengduan Mountains is a GD hotspot. In comparison, the Hd distribution pattern identifies more hotspots, with numerous cells with high Hd values distributed on the southeast edge of the QTP and the southern Hengduan Mountains. Three cells with high Hd values are distributed in the Qinling and Wuling Mountains. In summary, three Hd hotspots are detected: the southeast edge of the QTP, southern Hengduan Mountains, and Qinling Mountains. These areas are inferred to have been refugia during glacial cycle periods (Lei et al., 2015; Rahbek et al., 2019a); therefore, we propose that these montane regions have preserved a higher Hd than other regions (Qu et al., 2014; He et al., 2016). The hotspots of GD in vertebrates in China are consistent with hotspots of species diversity (Hu et al., 2021). Our results suggest a consistency between the GD and species diversity patterns in the MSC, especially the π pattern being more correlated with the species diversity pattern than Hd (Fjeldså, 2013; Wu et al., 2017).
Six of the nine cells with high π values (> 0.005) are located in the southern China zoogeographical region (Appendix Fig. S1A). In the early age of biogeography, Wallace (1876) has noticed a division across southern Yunnan, which has been supported convincingly by zoogeographical studies in different decades with multiple lines of evidence (Zhang, 1999; Holt et al., 2013; Dong et al., 2017). Several recent studies on avian phylogeny and taxonomy have revealed rich endemic and cryptic lineages in some passerine species within this region (Alström et al., 2016; Tritsch et al., 2017; Quintero and Jetz, 2018), advocating high phylo-lineage richness in the southern China zoogeographical region. Consistently, our results show an apparent difference of GD across the division and the high GD level in the southern China zoogeographical region.
There is no strong linear correlation between the GD parameters and the three geographical variables (latitude, longitude, and elevation). Whereas a poleward decrease of GD has been identified (Miraldo et al., 2016), our result indicates that patterns observed at a global scale may not apply to local scales, especially in the complicated topographical areas of the MSC (Lei, 2012; Rahbek et al., 2019b).
The "W" curves (both for π and Hd) indicate that GD changes in a rather complicated manner along the latitudinal gradient. We expect that the middle peak of GD (especially the Hd value) at 30° N might have been influenced by the mountain regions along this latitude. The unimodal pattern of GD along altRange also shows that the refugial function of MSC, most of these 1500 m altRange cells distributed in mountainous region. The topographical complexity of mountain regions can preserve more haplotypes than other regions nearby. Previous studies recorded a mid-elevation effect of species diversity, which means that the highest species richness is observed at a middle elevation in the MSC (Wu et al., 2017; Quintero and Jetz, 2018). In a recent study on passerine species diversity gradient, van Els et al. (2021) found that the highland area may have higher speciation rate than lowland, but the species diversity is higher in lowland, partly due to that the dispersal rate from highland to lowland is higher than opposite direction. In our study, the π and Hd are higher in highland area than lowland, maybe also due to the refugial effect within the high mountainous ranges which act as sky islands (He and Jiang, 2014; Lei et al., 2015). GD decrease with altitude and reach its lowest point ca. 2000 m, then increase and reach the highest point ca. 4000–4500 m. These different distribution patterns between GD and species richness also have been found in other taxa (Taberlet et al., 2012; Fan et al., 2018). We here propose that a possible cause for these patterns is the edge effect at different dimensions of biodiversity. At the species level, the species inhabiting low altitudes extend their distribution boundaries to mid-altitudes, and vice versa for high altitude-dwelling species, which could lead to high species numbers at mid-elevations (Cai et al., 2018). However, in the peripheral areas of a species' distribution, the population GD is expected to be lower than in the core area, as fewer genetic alleles are presumed to have been dispersed from the core area (Zhu et al., 2018b). Another possible factor may be insufficient sampling coverage. We only have sequences from 27 passerine species for analysis, while Wu et al.'s (2017) study on the species distribution pattern covers all the breeding birds of this area, 939 species of both passerines and non-passerines. In the future, sequences accumulation from more species may facilitate the study and clarify these findings on GD patterns along the geographical axis.
Correlation analysis reveals that the most correlated factor impacting π is the isothermality (BIO3) and precipitation seasonality (BIO15). The model selection results show that the best explanatory factors for π are NDVI, HII, and longitude. These results suggest that the temperature and precipitation-related variables, vegetation index and longitude gradient in together affect the π pattern. These results are consistent with the climate stability hypotheses for species richness (Gratton et al., 2017; Lawrence and Fraser, 2020), illustrating the importance of the stable climates in tropical and subtropical regions to the accumulation of genetic polymorphisms. A previous study has also found that the most correlated factors of the species richness of breeding birds in the Hengduan Mountains are related to temperature and precipitation (Wu et al., 2013).
Hd shows a significantly positive correlation with precipitation seasonality (BIO15). This result indicates that precipitation-related variables are important to Hd. Some researchers suggest that Hd is more related to recent population dynamics or adaptive changes to the environment (Martínez-Freiria et al., 2015; Zhu et al., 2018a). Our study proposes that the distribution pattern of Hd characterizes another important component of genetic diversity and may have been driven by different factors compared to those that drove π.
Human impacts at different scales may have contrasting effects on GD, and the HII may negatively impact species diversity patterns on a global scale (Millette et al., 2019). Our results at the local scale indicate this impact of human activities on the GD pattern.
In summary, correlation and model selection analyses reveal that GD is affected by multiple variables. The two measured indices of GD show variable results. Nucleotide diversity is driven by isothermality (BIO3), precipitation seasonality (BIO15), vegetation (NDVI), human influence (HII) and longitude. Hd is closely related to precipitation seasonality (BIO15) and altRange.
As a meta-analysis on genetic diversity at a comparatively narrow region, it is challenging to obtain a complete dataset sampling. Whereas we have collected and supplemented the dataset with the most efforts. Even so, we would suggest that the GD distribution patterns observed in this study should reserve a substantial confidence interval.
Our study has two prominent merits, which would shed lights on further and ongoing studies. First, the exploring efforts on the connection between the two dimensions of biodiversity – species richness vs genetic diversity at a regional scale. Previous studies have mainly concentrated at global or continental scale; however, this study focuses on a regional scale, in Mountains of Southwest China. It is rather necessary to carry out such studies on birds, as MSC is a global species richness hotspot preserving more than 900 species even in the future under the global climate change. Second, in our study the distribution pattern of π is in line with the species richness, demonstrating consistency of biodiversity at different dimensions within the MSC. The spatial pattern of Hd displays rather different modes, which may suggest different properties of the GD within this region.
Our results show that the Mountains of Southwest China harbor a highest GD similar to the species diversity, while the distribution patterns of π and Hd along the geographical axis are different. The pattern of GD distribution indicates a GD hotspot in southern China zoogeographical region, as well as a high/low divide, which is consistent with the boundary of the Central China and South China zoogeographical regions. In Mountains of Southwest China, latitudinal and longitudinal hypothesis could not explain the pattern well independently. Instead, a combination of environmental variables, such as temperature, precipitation, vegetation, human influence and topographical factors, can well explain the driving forces in shaping the current distribution patterns of genetic diversity.
The experiments comply with the current laws of China.
FL, GS and YC conceived the research project; CJ, YC, GS, FL and DZ collected the new samples; YC, GS and PF analyzed the data; DZ, YH and YC sequenced the new samples; YC, GS and FL led the writing, with contribution from YJ. All authors read and approved the final manuscript.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Many thanks to the editor and two anonymous reviewers for their very valuable comments and suggestions for improving our manuscript. This work was supported by National Natural Science Foundation of China (3213000355, 32070434, and 31900320), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA19050202) and the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0304, 2019QZKK0501).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.avrs.2022.100043.
|
Fischer, R.A., 1930. The Genetical Theory of Natural Selection. Oxford University Press, Oxford.
|
|
Heywood, V.H., Watson, R.T., 1995. Global Biodiversity Assessment. Cambridge University Press, Cambridge.
|
|
Lei, F.M., 2012. Global endemism needs spatial integration. Science 335, 284-285.
|
|
Millette, K.L., Fugere, V., Debyser, C., Greiner, A., Chain, F.J.J., González, A., 2019. No consistent effects of humans on animal genetic diversity worldwide. Ecol. Lett. 23, 55-67.
|
|
Mittermeier, R.A., Turner, W.R., Larsen, F.W., Brook, T.M., Gascon, C., 2011. Global Biodiversity Conservation: the Critical Role of Hotspots. Springer, Heidelberg.
|
|
Nei, M., 1987. Molecular Evolutionary Genetics. Colombia University Press, New York.
|
|
Wang, W.J., Dai, C.Y., Mckay, B.D., Zhao, N., Li, S.H., Lei, F.M., 2013. Microsatellites underestimate genetic divergence in the Green-backed Tit (Parus monticolus). Avian Res. 4, 144-154.
|
|
Wickham, H., 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York.
|
|
Zhang, R.Z., 1999. China Animal Geography. Science Press, Beijing.
|
| Spearman rho | π | Hd |
| BIO1 | 0.088 | -0.195 |
| BIO3 | 0.315* | 0.199 |
| BIO4 | -0.196 | -0.057 |
| BIO7 | -0.135 | 0.094 |
| BIO12 | -0.034 | -0.207 |
| BIO15 | 0.279* | 0.217* |
| NDVI | -0.093 | -0.234 |
| EVI | 0.084 | 0.243 |
| HII | 0.027 | -0.143 |
| Altitude | 0.021 | 0.236 |
| altRange | -0.100 | -0.120 |
| Longitude | -0.160 | -0.218 |
| Latitude | -0.111 | 0.098 |
| π, nucleotide diversity, Hd, haplotype diversity. *P < 0.05, **P < 0.01. | ||
| Index | Selection variables |
| π | NDVI, HII, longitude |
| Hd | BIO15, altRange |