| Citation: | Michael S. Lukubwe, Daniel Velarde-Garcéz, Fernando Sequeira, Susana Lopes, Adrian J.F.K. Craig, Vanessa A. Mata. 2024: Feeding ecology and interactions with mammal hosts in a symbiotic genus of birds (Buphagus spp.) in Namibia. Avian Research, 15(1): 100200. DOI: 10.1016/j.avrs.2024.100200 |
As the sole obligate symbiotic birds in Africa, oxpeckers offer a unique model for studying symbiotic relationships. Due to the multitrophic level they occupy and the context dependent foraging behavior they exhibit, the type of symbiotic relationship can be variable. In addition to providing a cleaning service to the host by removing ticks, oxpeckers frequently feed on blood, mucus, and saliva, inflicting potential damage on the host. Here, we used DNA metabarcoding on faecal samples to analyze the taxonomic composition of the trophic interactions of the Yellow-billed Oxpecker (Buphagus africanus) and Red-billed Oxpecker (B. erythrorhynchus) in northeastern Namibia. In contrast to conventional methods, DNA metabarcoding allows for a detailed identification of dietary resources encompassing both mammal hosts and consumed arthropods within the same samples. With this information, we examined differences in the diet composition between oxpecker species and localities, as well as the co-occurrence between host and arthropod species. Our findings revealed that oxpeckers predominantly source their diet from mammals, ticks, and flies; however, ticks and flies rarely co-occur in the diet of an individual. We observed variability among individuals in their feeding ecology, which is strongly correlated with locality and, to a lesser extent, with the mammal host. We noted a high degree of mobility between hosts within relatively short periods, with 32% of the samples showing traces of at least two mammal hosts. This study illustrates the dynamic foraging behavior of these specialized symbiotic birds, shedding light on their potential role in pest control services and disease transmission.
Mutualism and parasitism are usually presented as mutually-exclusive and discrete categories, albeit it is becoming increasingly clear that the net gain of an interaction follows a continuous gradient. In many cases, it does not show a clear division between the different types of symbiosis (Leung and Poulin, 2008). In broad terms, mutualistic interactions are characterised by net benefits to both interacting participants, and parasitism by harm to one of the participants while the other benefits. This distinction becomes problematic in mutualistic relationships in which the service presents a high cost for one of the participants, or in parasitic interactions with low levels of harm, and especially when there is high variation in the behavior within the population providing the service (Cheney and Côté, 2005; Leung and Poulin, 2008). For example, in cleaning fishes (e.g., Elacatinus evelynae, Labroides dimidiatus) there is evidence of the influence of personality and resource availability on the frequency of ‘cheating’ behavior during mutualistic interactions (Cheney and Côté, 2005; Wilson et al., 2014). Extensive research in behavioral ecology has explored mutualism and parasitism, often focusing on game theory and its implications for fitness. Nevertheless, even though symbiotic relationships usually arise from feeding interactions, there has been limited investigation into the impact of trophic complexity on such relationships (Hoeksema and Bruna, 2000). Since many mutualist species engage in multitrophic interactions, it becomes challenging to produce detailed descriptions of the taxonomic diversity in their diet. For instance, Broadstripe Gobies (Elacatinus prochilos) can include sponges and polyps to their diet mainly composed from isopod larvae from the skin of other fishes (Arnal and Côte, 2000).
Oxpeckers serve as a unique model for the study of symbiotic relationships, exhibiting an extreme degree of specialization to the extent of being the only obligate symbiotic bird in Africa (Acquaviva and Junior, 2022). There is little consensus among academics regarding the type of symbiotic interaction they engage in. Aside from providing a cleaning service to the host by removing ticks, they frequently feed on blood, mucus, and saliva, which can be quite costly for the host (Plantan et al., 2013). By feeding on ticks, oxpeckers reduce the tick load on host animals, preventing issues such as blood loss, appetite suppression, tick toxicosis, and skin infections from bacterial and protozoan diseases (Plantan et al., 2013; Mashebe et al., 2014). Nevertheless, oxpeckers usually feed on engorged ticks, which have already drained blood from the host and transferred any potential diseases to the host animal (Plantan et al., 2013).
The oxpeckers can also inflict injuries on host animals and keep these wounds open, especially during times of low tick availability (Samish and Rehacek, 1999; Abakpa et al., 2023). Furthermore, anecdotal evidence suggests that feeding on open wounds of wild ungulates by oxpeckers facilitates the transmission of diseases, both within species and between wild and domestic populations (Han et al., 2022; Kioko et al., 2022). Since oxpeckers feed on their host mammals and the ticks and flies cohabiting with them, they occupy a multitrophic level, adding a layer of complexity to their mutualistic behavior. Unlike other mutualistic species, where their services may be impacted by resources outside the symbiotic relationship, all of the primary feeding resources for oxpeckers come from within their symbiotic relationship. As previously suggested, it is expected that the dynamics of mutualistic interactions are shaped by the presence of additional trophic levels (Hoeksema and Bruna, 2000).
The close proximity of oxpeckers to human populations, their unusual foraging behavior, and their ambiguous position as parasites and mutualists, have led to numerous observational and experimental studies on the relationship between oxpeckers and hosts or ticks. Past research on the symbiotic interactions of oxpeckers reveals that they use a wide variety of hosts, from Domestic Cattle (Bos taurus) to large species (e.g., Hippopotamus amphibius, Ceratotherium simum), medium-sized species (e.g., Equus quagga, Tragelaphus strepsiceros), and small species (e.g., Aepyceros melampus) of wild ungulates (Dale, 1992; Mooring and Mundy, 1996; Ndlovu and Combrink, 2015). They appear to be highly specialized on ticks, showing some degree of preference for certain species, such as Blue Ticks (Boophilus decoloratus) and Brown Ear Ticks (Rhipicephalus appendiculatus) (Robertson and Jarvis, 2000; Weeks, 2000).
Host selection by oxpeckers has been repeatedly studied across their wide geographic range (e.g., Tarakini et al., 2017; Lukubwe et al., 2023). Evidence suggests a preference for larger mammal species (Ward and Robertson, 2017; Gagnon et al., 2020); however, since there is competition among oxpeckers for the hosts (Peron et al., 2019), selection varies across their geographical range, depending on the mammal species available and whether both species of oxpeckers co-occur (Ndlovu and Combrink, 2015). On the other hand, significantly less emphasis has been placed on the selection of ectoparasites that oxpeckers feed on (e.g., Bezuidenhout and Stutterheim, 1980; Stutterheim et al., 1988). Studies of the trophic ecology of oxpeckers are limited to direct observations of their foraging behavior and stomach content analysis (reviewed in Weeks, 2000). While these approaches are helpful, they either estimate the proportion of time invested in different host species or the relative consumption of arthropod species. This is because the analysis of stomach content does not allow for the identification of the host on which the individuals were feeding (e.g., Moreau, 1933; Bezuidenhout and Stutterheim, 1980), while studies that register the feeding frequency in different mammal species are unable to accurately address the taxonomic identity of the arthropods (e.g., Stutterheim, 1981; Ndlovu and Combrink, 2015). Closing this knowledge gap is crucial for comprehending the complex foraging behavior of oxpeckers and their influence within the multi-level trophic network.
Here, we used DNA metabarcoding on fecal samples to analyze the taxonomic composition of the trophic interactions of the Yellow-billed (Buphagus africanus) and Red-billed Oxpecker (Buphagus erythrorhynchus) in northeastern Namibia. The identification of dietary resources through DNA metabarcoding allows for high taxonomic detail in the identification of mammalian hosts and consumed arthropods within the same samples. With this information, we tested for differences in the diet composition between oxpecker species and localities, as well as the co-occurrence between dyads of hosts, arthropods, and host-arthropods. As suggested by the literature (Plantan et al., 2013; Acquaviva and Junior, 2022), we expected a high degree of specialization in hosts and tick species. Although there is evidence that Yellow-billed Oxpeckers tend to prefer larger-sized ungulates than Red-billed Oxpeckers (Mikula et al., 2018), we expected to find additional differences in the taxonomic identity of the foraged arthropods. Finally, we expected to find dyads of species that co-occur both lower and higher than expected by chance, most likely as a result of the spatial distribution of the mammals and the parasitic relationship between mammals and arthropods.
The Salambala Conservancy has a population of approximately 9193 people (NACSO, 2023). The conservancy covers an area of 9300 ha, with 18 villages and 290 cattle posts (NACSO, 2023). It is home to a diverse range of wildlife, including elephants, lions, and giraffes. The local community relies heavily on the conservancy for their livelihood, with many engaged in tourism and conservation efforts. The area is largely woodland (59%) and grassland (41%) (Mendelsohn et al., 1997). The conservancy's northern half is dominated by mopane and Kalahari woodlands, while the southern half is dominated by grasslands (Mendelsohn et al., 1997). Annual rainfall is 650–700 mm, with 95% of the rain falling between October and April (Mendelsohn et al., 1997; Krug, 2017), and most of it occurs during the rainy season (November to April). The coldest temperatures are registered in July, while the highest temperatures occur from September to November (Mendelsohn et al., 1997). Salambala is quite flat, with an elevation between 928 m and 983 m above sea level (Mendelsohn et al., 1997). Floodplains dominate riverine drainage flooding during heavy rain.
Samples were collected at Izumba Village (17°52′40″ S, 24°44′00″ E), Chobe River Camp (17°53′53″ S, 24°43′43″ E), and at Ioma Village (17°53′13″ S, 24°24′29″ E) in the Salambala Conservancy (Fig. 1). The study sites were selected by searching for the regular presence of both oxpecker species during an early field reconnaissance from 2019 to 2022.
Mist nets were placed at livestock enclosures to capture the birds. The mist nets remained open every day from 5:00 to 10:00 a.m. while the cattle were in an existing livestock enclosure built using wire mesh and steel poles. We chose this enclosure to avoid nets becoming entangled and teared in the process, which is a problem when setting up nets in traditional thorn-bush enclosures. Nets were placed outside the fence in two lines, each measuring 12 × 24 m long, and running side by side at a height of 5.5 m and 3.5 m. Thus, any birds escaping the first line would fly into the second. Two 25-m ropes, bolstered by metal cans, kept the cattle in the enclosure until two research assistants entered to drive the cattle, and thus the birds, towards the nets. Additionally, we used playback recordings of oxpeckers to improve the chances of attracting birds to the nets. Once captured, birds emitted distress calls, which lured more oxpeckers to the nets. Captured birds were left in the nets for approximately 10 min, resulting in the capture of additional oxpeckers. Once the oxpeckers were captured, the nets were dropped, and the birds were removed. The oxpeckers were then held in hanging cloth bags out of the sun until they were processed. We placed filter paper in the bottom of every clean cloth bag where the birds were kept prior to handling. We removed the paper after the birds had defecated and placed the samples in individual tubes containing 70% ethanol. The samples were first air-dried using a centrifuge and then sent for genetic analysis.
DNA was extracted from samples using the E.Z.N.A. Tissue Kit (Omega), while following the manufacturer's recommendations. Due to the lack of proper and solid droppings, the piece of paper containing feces remains was added to the buffer with sterile tweezers. DNA extractions were done in batches of 23 samples plus one negative control in which no sample was added. The diet of oxpeckers was assessed by amplifying a small fragment (178 bp) of the mitochondrial cytochrome c oxidase I gene (COI) using the primers fwh1 (Vamos et al., 2017) modified with Illumina overhang adapter sequences. Although these primers were initially designed for the amplification of freshwater invertebrates, their high degeneracy level allows them to amplify a wide range of other taxa, including most vertebrates. This, along with the small size of the targeted region, makes this primerset ideal for the amplification of degraded DNA resulting from the digestion of both the invertebrate prey and blood and skin of the vertebrate host of oxpeckers. Due to the primer's capacity in amplifying oxpeckers' DNA, a blocking primer was designed (Buph-erythro_fwhF1-block: 5′–CAAAGATATCGGCACTCTCTACCTAATCTTC[SpC3]–3′) to reduce this amplification by aligning Fwh1 primers with existing COI sequences of both Buphagus species in BOLD and NCBI. The designed primer perfectly matched the available sequence of B. erythrorhynchus, our most abundant species, but showed two mismatches to B. africanus which could slightly reduce its blocking efficiency, but should nonetheless have some effect. PCR reactions consisted of 5 μL of Qiagen Multiplex Master Mix, 0.25 μL of each 10 μM primer, 0.38 μL of 100 nM blocking primer, 2.13 μL of water, and 2 μL of DNA extract. Cycling conditions consisted in a 15 min period at 95 ℃, 40 cycles of 30 s denaturation at 95 ℃, 30 s annealing at 50 ℃, and 30 s extension at 72 ℃, and a final extension period of 10 min at 72 ℃. PCR success was checked in a stained agarose gel and for samples that were not successfully amplified, PCR was repeated using 3 μL of DNA extract and 45 cycles of amplification. PCR products were diluted 1:3 or 1:4 depending on band intensity and went through a second PCR reaction to incorporate 7 bp long indexes and P5 + P7 Illumina adaptors. PCR reactions consisted in 7 μL of KAPA HiFi HotStart ReadyMix (Rocher), 0.7 μL of each indexing primer, 2.8 μL of water, and 2.8 μL of diluted PCR product. Cycling conditions consisted in a 3 min period at 95 ℃, 10 cycles of 30 s denaturation at 95 ℃, 30 s annealing at 55 ℃, and 30 s extension at 72 ℃, and a final extension period of 10 min at 72 ℃. Indexed PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter) with a ratio of 1:0.8, and subsequently quantified using Epoch Microplate Spectrophotometer (Agilent Technologies, Santa Clara, USA) and diluted to 15 nM. Libraries were pooled equimolarly and the pool was quantified with qPCR (KAPA Library Quant Kit qPCR Mix, Rocher) and diluted to 4 nM. Finally, the pool was sequenced in an Illumina MiSeq v2 kit (500 cycles) for a target depth of 60k paired reads per sample.
Paired reads were merged using ‘illuminapairedend’ of Obitools (Boyer et al., 2016), while removal of primer sequences and tagging of reads with sample information was done with ‘ngsfilter’. Reads were then dereplicated per sample using the command ‘obiuniq’, and singletons of each sample were removed with the command ‘obigrep’. Afterwards, reads of each sample were merged and denoised with the command ‘--cluster_unoise’ of VSEARCH (Rognes et al., 2016). Potential chimaeras were removed using the command ‘--uchime3_denovo’ and the remaining zOTUs were clustered at 99% similarity using ‘--cluster_size’. Merged reads were then mapped to the retained OTUs using the command ‘--usearch_global’ with an identity level of 99%. To reduce the number of PCR artefacts, sequencing errors, and nuclear copies of the mitochondria, LULU (Frøslev et al., 2017) was used to merge similar OTUs (identity >84%) with high co-occurrence levels (>95% of samples). Resulting OTUs were identified with BOLDigger (Buchner and Leese, 2020) and manually inspected to validate their identity. OTUs were classified as ‘diet’ or ‘not diet’ depending on their identity, with non-target taxa like fungi, bacteria, parasites, acari, among other non-arthropod or non-mammalian taxa, being classified as ‘not diet’. Samples with less than 100 reads belonging to ‘diet’ items were considered to have failed and were removed. To remove potential lab contaminations, the number of reads observed per OTU present in extraction and PCR blanks was subtracted from the respective samples associated with each extraction and PCR blank. Finally, from each sample we further removed all taxa representing less than 1% of the total number of dietary reads of that sample (Mata et al., 2019).
Data analysis was conducted using R v4.3.0 (R core Team, 2022) considering the presence/absence of prey items on samples at the Operational Taxonomic Unit (OTU) level. We used OTUs instead of species as the most resolved taxonomic level, since some taxa were identified only to family or order and then clustered into groups. The asymptotic richness was estimated using rarefaction curves based on Hill numbers with the function ‘iNEXT’ of the package ‘iNEXT’ (Hsieh et al., 2016).
Differences in the dietary composition between oxpecker species and localities were assessed using a Permutational Multivariate Analysis of Variance (PERMANOVA) with 10,000 permutations, using the function adonis2 from the package vegan (Dixon, 2003) based on a Jaccard dissimilarity matrix. Multivariate homogeneity of group dispersion was assessed using the function ‘betadisper’ of the package ‘vegan’, and then it was tested if distance to centroid was significantly different between categories with the functions ‘anova’ and ‘TukeyHSD’ of the package ‘stats’ (R core Team, 2022). To identify the prey items that contributed the most to differences in dietary composition, the function ‘simper’ of the package ‘vegan’ was used with 10,000 permutations. We used a Generalized Linear Model (GLM) to assess the differences in individual diet richness and phylogenetic specialization of mammal hosts across oxpecker species and localities. Poisson distribution was used for diet richness and beta distribution for phylogenetic species. Phylogenetic specialization was calculated using the specificity index (STD) described in Poulin and Mouillot (2003) and then transformed to frequencies by diving the STD of each sample by four, which is the upper limit of the index. To evaluate if co-occurrence between dietary resources was higher or lower than expected by chance, we used the probabilistic model presented in Veech (2013). For data visualisation, we employed a Correspondence Analysis for the significant variables according to the PERMANOVA. This method enables us to reconstruct the distances between samples and their associated prey items. To visualise the co-occurrence patterns among prey items, we employed Non-Metric Multidimensional Scaling (NMDS). NMDS creates a two-dimensional map illustrating the associations between prey items based on their shared presence in samples. For all analysis, statistical significance was considered at an alpha value of 0.05.
After completing laboratory and bioinformatic procedures, we successfully obtained diet composition data from a total of 66 samples derived from the initial capture of 83 individuals. From this, 51 samples belonged to the Red-billed Oxpecker and 15 to the Yellow-billed Oxpecker. Average number of reads ± standard deviation of mammal OTUs by sample was 16,024 ± 17,707, and for arthropod OTUs by sample 3156 ± 5789.
We identified 19 OTUs, of which 6 were mammals and 13 arthropods (see Table 1). From the obtained OTUs 14 were identified to species level. One OTU was identified to family level, belonging to Ixodidae. Four OTUs were identified to order level, all belonging to Diptera. Estimated sample coverage of OTUS was 0.97, and asymptotic richness 21.46 (Confidence interval of 95%: 19–38). Samples showed an average richness of 2.85 ± 1.35 OTUs. At least one mammal species was identified in 95% of the samples and at least two mammals species in 32%, while arthropods were present in 83% of the samples. From the total of samples, 16% had only traces of mammal in the diet, suggesting that they only fed directly on the mucus or blood; and 41% of the samples had mammal DNA but no tick DNA, suggesting that either secondary consumption occurred via blood-feeding flies, or the detection corresponds to direct feeding from the host. The most common OTU was Bos taurus (prevalence of 92%), followed by Rhipicephalus decoloratus (42%) and Diptera 4 (38%). For families, Bovidae (94%) showed the highest prevalence, followed by Ixodidae (59%) and Giraffidae (11%). The most common orders were Artiodactyla (94%), Ixodida (59%) and Diptera (42%; see Fig. 2).
| Phylum | Class | Order | Family | OTU | Oxpecker species | Localities | |||||||||
| Red-billed Oxpecker | Yellow-billed Oxpecker | Izumba Village | Ioma Village | Chobe River Lodge | |||||||||||
| Total=51 | Total=15 | Total=30 | Total=28 | Total=4 | |||||||||||
| N | % | N | % | N | % | N | % | N | % | ||||||
| Arthropoda | Arachnida | Araneae | Cheiracanthiidae | Cheiracanthium 1 | 0 | 0.00% | 1 | 6.67% | 0 | 0.00% | 0 | 0.00% | 1 | 25.00% | |
| Ixodida | Ixodidae | Amblyomma variegatum | 9 | 17.65% | 0 | 0.00% | 1 | 3.33% | 8 | 28.57% | 0 | 0.00% | |||
| Hyalomma truncatum | 1 | 1.96% | 1 | 6.67% | 1 | 3.33% | 0 | 0.00% | 0 | 0.00% | |||||
| Ixodidae 1 | 17 | 33.33% | 1 | 6.67% | 4 | 13.33% | 13 | 46.43% | 1 | 25.00% | |||||
| Rhipicephalus decoloratus | 23 | 45.10% | 5 | 33.33% | 6 | 20.00% | 19 | 67.86% | 3 | 75.00% | |||||
| Rhipicephalus evertsi | 11 | 21.57% | 4 | 26.67% | 5 | 16.67% | 8 | 28.57% | 2 | 50.00% | |||||
| Rhipicephalus zambeziensis | 2 | 3.92% | 0 | 0.00% | 1 | 3.33% | 1 | 3.57% | 0 | 0.00% | |||||
| Insecta | Coleontera | Tenebrionidae | Zophobas atratus | 1 | 1.96% | 1 | 6.67% | 2 | 6.67% | 0 | 0.00% | 0 | 0.00% | ||
| Diptera | Ceratopogonidae | Culicoides leucostictus | 1 | 1.96% | 0 | 0.00% | 0 | 0.00% | 1 | 3.57% | 0 | 0.00% | |||
| Not identified | Diptera 1 | 1 | 1.96% | 0 | 0.00% | 0 | 0.00% | 1 | 3.57% | 0 | 0.00% | ||||
| Diptera 2 | 1 | 1.96% | 0 | 0.00% | 1 | 3.33% | 0 | 0.00% | 0 | 0.00% | |||||
| Diptera 3 | 1 | 1.96% | 0 | 0.00% | 0 | 0.00% | 1 | 3.57% | 0 | 0.00% | |||||
| Diptera 4 | 16 | 31.37% | 9 | 60.00% | 19 | 63.33% | 2 | 7.14% | 1 | 25.00% | |||||
| Chordata | Mammalia | Artiodactyla | Bovidae | Aepyceros melampus | 2 | 3.92% | 0 | 0.00% | 0 | 0.00% | 2 | 7.14% | 0 | 0.00% | |
| Bos taurus | 47 | 92.16% | 14 | 93.33% | 25 | 83.33% | 28 | 100.00% | 4 | 100.00% | |||||
| Hippotragus niger | 2 | 3.92% | 0 | 0.00% | 2 | 6.67% | 0 | 0.00% | 0 | 0.00% | |||||
| Syncerus caffer | 2 | 3.92% | 2 | 13.33% | 3 | 10.00% | 0 | 0.00% | 1 | 25.00% | |||||
| Giraffidae | Giraffa camelopardalis | 7 | 13.73% | 0 | 0.00% | 2 | 6.67% | 5 | 17.86% | 0 | 0.00% | ||||
| Perissodactyla | Equidae | Equus burchellii | 6 | 11.76% | 0 | 0.00% | 6 | 20.00% | 0 | 0.00% | 0 | 0.00% | |||
The analysis of variance (ANOVA) for the distance to the centroid indicated a significant variation in the dispersion of dissimilarity values in diet composition among different oxpeckers’ species (df = 1, Sum-Sq = 0.12193, Mean-Sq = 0.121928, F = 4.5504, p = 0.03675), but not among localities (df = 2, Sum-Sq = 0.05727, Mean-Sq = 0.028635, F = 0.954, p = 0.3907). PERMANOVA did not reveal differences between the two oxpecker species (df = 1, pseudo-F = 2.58, R 2 = 0.03, p = 0.05), but it supported differences in dietary composition between localities (df = 3, pseudo-F = 5.42, R 2 = 0.20, p = 0.0001; see Fig. 3). According to SIMPER, four arthropod species explained the dietary variance among localities. Tick species (Rhipicephalus decoloratus, Ixodidae 1 and Amblyomma variegatum) were more common in Ioma Village, while Diptera 4 was mostly consumed in Izumba Village. Depending on the tick species, the prevalence in the diet was between 3.4 and 8.6 times higher in Ioma Village. In Izumba, oxpeckers focused on flies, as suggested by the presence of Diptera 4 in 65% of the samples, while only 35% of the samples had any of the six tick species detected in this study. In contrast, in Ioma, Diptera 4 was not among the most common prey items, being present in only 7% of the samples.
The mean richness of OTUs was slightly higher for Red-billed Oxpeckers (2.94 ± 1.44) compared to Yellow-billed Oxpeckers (2.53 ± 0.91), although this difference was not significant (df = 1, p = 0.4043). A similar non-significant pattern was observed when decomposing the richness into mammal OTUs (df = 1, p = 0.4795) and arthropod OTUs (df = 1, p = 0.6243). For mammal OTUs, Yellow-billed Oxpeckers had a mean richness of 1.29 ± 0.64, while Red-billed Oxpeckers had a mean richness of 1.06 ± 0.45. For arthropod OTUs, Yellow-billed Oxpeckers had a mean richness of 1.64 ± 1.23, and Red-billed Oxpeckers had a mean richness of 1.46 ± 0.74. Although not significant (df = 1, p = 0.2200), phylogenetic specialization with mammal hosts was higher for Red-billed Oxpeckers (1.59 ± 1.01) than for Yellow-billed Oxpeckers (1.14 ± 0.36). Similarly, no significant difference was found across localities, either for diet richness (df = 2, p = 0.4408) or phylogenetic specialization (df = 2, p = 0.5842).
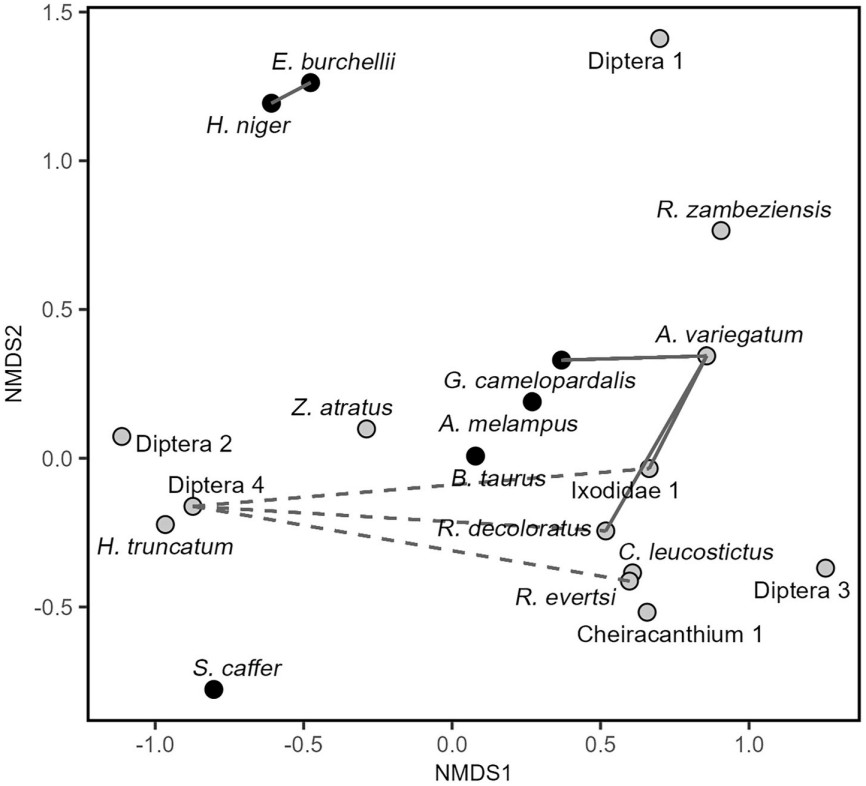
The analysis of co-occurrence between dietary resources supported five dyads of species that co-occur more than expected and three dyads that co-occur less often than expected (see Fig. 4). The host-tick dyad Giraffa camelopardalis–Amblyomma variegatum (p = 0.047), the host-host dyad Hippotragus niger–Equus burchellii (p = 0.006) and the tick-tick dyad of Ixodidae 1-Amblyomma variegatum (p = 0.009), Rhipicephalus decoloratus–Amblyomma variegatum (p = 0.026) and Rhipicephalus decoloratus–Ixodidae 1 (p <0.001), co-occurred more often than expected. Otherwise, Diptera sp.4 co-occurred less than expected by chance with the tick species Ixodidae 1 (p = 0.005), Rhipicephalus decoloratus (p = 0.016) and Rhipicephalus evertsi (p = 0.023). For example, individuals that fed on giraffes were more likely to prey on Amblyomma variegatum than expected, with 43% of the oxpeckers that fed on giraffes preying on this species, in contrast to the 13% prevalence of A. variegatum in all samples.
Our study highlights the utility of employing high-throughput sequencing in fecal samples to reconstruct the trophic ecology of a vertebrate symbiont and, consequently, to infer the heterogeneity of strategies employed within their populations. For symbiotic interactions in which a species feeds on both mammalian hosts and their ectoparasites, DNA metabarcoding allows us to simultaneously monitor both levels of their trophic ecology. As a result, we generated a description of the interactions of oxpeckers with their mammal hosts and the arthropods on which the oxpeckers feed. As previously suggested (Plantan et al., 2013), we found that the diet of oxpeckers is mostly sourced from mammals and ticks. However, flies and mosquitoes, which are reported infrequently in the literature (to our knowledge only in van Someren, 1951; Stutterheim, 1981), appear to be frequently consumed in these Namibian oxpecker populations. We also observed that, aside from ticks and flies, oxpeckers can feed on other arthropods, such as beetles or spiders, albeit this is rare. The narrow diet of oxpecker populations and the low richness of the individual diet support the notion that they are highly specialized on large mammals and the community of arthropods associated with these. Nevertheless, we observed some variability among individuals in their feeding ecology, strongly correlated with the locality and, to a lesser extent, with the mammal host.
Although it was previously assumed that oxpeckers mainly fed on ticks (Stutterheim, 1981; Koenig, 1997), it has progressively become clear that blood feeding occurs in equal or higher proportion (Weeks, 2000; Plantan et al., 2013). In this study, mammals appear to be the most consistent food source, due to the dominance of Bos taurus in the diet. However, DNA metabarcoding does not allow quantification of the percentage of mammal detections due to secondary consumption, since feeding directly on the mucus or blood can only be inferred for samples with solely traces of mammals. This limitation of DNA metabarcoding methods prevents us from inferring about the type of symbiotic relationship with their hosts. However, the well-resolved diet composition is useful for detecting subtle dietary differences and provides a multitrophic perspective on understanding the effects that mammal hosts have on their arthropod feeding strategies.
The spatial variation in the intensity of tick consumption that we found aligns with earlier studies suggesting that foraging for blood or ticks by oxpeckers is context-dependent (Plantan et al., 2013). However, the unexpectedly high consumption of Diptera and low consumption of Ixodidae in one locality could also suggest that the dependency of oxpeckers on ticks has been overestimated. Our findings show that the prevailing foraging strategy can vary between populations, likely influenced by differences in resource availability. There are some environmental differences across localities that probably contribute to the shift in diet composition. Precipitation and vegetation cover are higher on the east side of the Sambalala Conservancy (NACSO, 2022), where Izumba Village is located, than on the west side, where Ioma Village is situated. Due to this, and the fact that the east side of the conservancy is not fenced to allow the movement of mammals with Botswana, the surroundings of Izumba Village have a richer community of mammals, which is reflected in the diet of the oxpeckers. Additionally, the sampling point in Ioma Village is surrounded by a higher human population density than Izumba, which could lead to more intensive livestock activity and a higher density of ticks, possibly explaining the more frequent ingestion of ticks in this locality.
Only one association between a mammal host and a tick species was found. Despite past evidence suggesting that A. variegatum is more common in cattle than in wild mammals (Norval et al., 1994), we found that it was more likely to be present in oxpeckers that fed in giraffes. It is possible that in our study area, A. variegatum occurs more often in giraffes or is located in more accessible parts of their bodies. Additionally, the low co-occurrence between Diptera 4 and the three species of ticks suggests that there is either low spatial overlap between the arthropod species, oxpeckers display short-term specialization in a foraging strategy, or a combination of both. The co-occurrence between burchell's zebra and sable antelope is probably due to the spatial proximity of their grazing areas. In Mozambique it has been observed that during the wet season, when resources are abundant, the core area of both mammals overlap, and they are occasionally observed foraging together (Macandza, 2009). Due to the high co-occurrence between sable antelope and burchell's zebra, and with 32% of the samples having traces of at least two mammal host species, we infer high mobility across host species within a relatively short period. High-throughput analysis of bird's feces typically detects DNA ingested within the last 30 min to 4 h before the sample was collected (Oehm et al., 2011). Furthermore, of the 18 samples containing DNA of wild mammals, all except two also had traces of cattle. This illustrates the dynamic foraging behavior of oxpeckers and highlights their potential contribution to disease transmission between cattle and wild herbivore mammals.
Although no significant difference in diet was observed between oxpecker species, this is likely influenced by the low sample size for Yellow-billed Oxpeckers. Red-billed Oxpeckers showed a slightly higher diet richness of arthropods and mammals. Yellow-billed Oxpeckers only used cattle and buffalo as hosts, which is consistent with the findings of a previous study in the region that buffaloes are their most frequently used host (Ward and Robertson, 2017). Given that sample collection for this study was conducted in settlements with a high density of cattle, we cannot be certain that oxpeckers prefer cattle to wild ungulates. In future studies, research into the trophic ecology of oxpeckers should aim to distribute sampling locations between settlements and core protected areas. This approach will help to better understand the variability in host selection and its relationship to the consumption of arthtopods. Nonetheless, our study contributes to understanding the foraging patterns of oxpeckers in livestock areas, providing insights into their foraging strategies, pest-control services, and mobility. This will help us estimate the possible contributions of oxpeckers to cattle pest management services and their role in wildlife conservation. Although oxpeckers are obligate symbionts of mammals, they exhibit some degree of plasticity in their foraging strategy, covering a wide range of mammal species and showing context-dependent specialization on different arthropods. It is probable that the frequency of feeding on blood directly from a wound varies across localities due to changes in resource availability, similar to the intensity of tick consumption. Further research on the trophic ecology of oxpeckers can enhance our understanding of the evolution of mutualism in vertebrates or the subtle differences between mutualism and parasitism.
This study was approved by the Rhodes University Ethics Committee-Animal Research Ethics Committee (RU-AREC, Approval Number: 2019-0746-823 & 2019-1194-3186). A research permit was obtained from the National Commission on Research Science and Technology of Namibia (NCRST, Permit Number: RPIV00642019) and the Namibia Chamber of Environment (NCE, certificate no: RCIV00042018 from 2018 to 2024).
Michael S. Lukubwe: Conceptualization, Investigation, Methodology, Project administration, Writing – original draft. Daniel Velarde-Garcéz: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. Fernando Sequeira: Conceptualization, Writing – review & editing. Susana Lopes: Formal analysis, Methodology, Writing – review & editing. Adrian J.F.K. Craig: Conceptualization, Writing – review & editing. Vanessa A. Mata: Data curation, Methodology, Supervision, Writing – original draft.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
We are grateful to Frieda Ashipala and Block Roland for their assistance in delivering the samples to University of Porto (CIBIO), Portugal.
|
Abakpa, S.A.V., Adeosun, K.E., Akintunde, G.O., Egbetade, A.O., Olasoju, M.I., Alamu, A.O., et al., 2023. Effect of oxpeckers' interactions on wounds healing process in calves at Federal University of Agriculture cattle production farm, Abeokuta, Southwest Nigeria. J. Sust. Vet. All. Sci. 4, 96–100.
|
|
Acquaviva, J., Guimarães Junior, P.R., 2022. Generalist diets allows opportunism
behavior to engage in cleaning interactions among birds and large mammals. SciELO
Preprints.
|
|
Bezuidenhout, J.D., Stutterheim, C.J., 1980. A critical evaluation of the role played by the red-billed oxpecker Buphagus erythrorhynchus in the biological control of ticks. Onderstepoort J. Vet. Res. 47, 51–75.
|
|
Buchner, D., Leese, F., 2020. BOLDigger – a Python package to identify and organise sequences with the Barcode of Life Data systems. Metab. Metag. 4, e53535.
|
|
Hsieh, T.C., Ma, K.H., Chao, A., 2016. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Method. Ecol. Evol. 7, 1451–1456.
|
|
Kioko, J., Moore, S., Moshofsky, K., Nonnamaker, A., Ebanietti, B., Thompson, K., et al.,
2022. Characterizing elephant-livestock interactions using a social-ecological
approach. In: Kiffner, C., Bond, M.L., Lee, D.E. (Eds.), Tarangire: Human-Wildlife
Coexistence in a Fragmented Ecosystem, Ecological Studies. Springer International
Publishing, Cham, pp. 277–294.
|
|
Leung, T., Poulin, R., 2008. Parasitism, commensalism, and mutualism: exploring the many shades of symbioses. Vie Milieu , 107–115.
|
|
Lukubwe, M.S., Craig, A.J.F.K., Manyangadze, T., 2023. Oxpecker host-selection in the Salambala Conservancy, northeastern Namibia. Afr. J. Wildl. Res. 53, 166–176.
|
|
Macandza, V.A., 2009. Resource partitioning between low-density and high-density
grazers: sable antelope, zebra and buffalo. University of the Witwatersrand,
Johannesburg. Doctoral Thesis.
|
|
Mashebe, P., Lyaku, J.R., Mausse, F., 2014. Occurrence of ticks and tick-borne diseases of livestock in Zambezi Region: a review. J. Agr. Sci. 6, 142.
|
|
Mendelsohn, J., Robets, C., Hines, C., 1997. An Environmental Profile and Atlas of
Caprivi. Directorate of Environmental Affairs, Windhoek, Namibia.
|
|
Moreau, R.E., 1933. The food of the Red-billed oxpecker, Buphagus erythrorhynchus (Stanley). Bull. Entomol. Res. 24, 325–335.
|
|
NACSO (Namibia Association of CBNRM Support Organisation), 2022. Salambala:
Climate and Vegetation Report. NACSO, Windhoek, Namibia.
|
|
NACSO (Namibia Association of CBNRM Support Organisation), 2023. Salambala
Conservancy. NACSO, Windhoek, Namibia.
|
|
Ndlovu, M., Combrink, L., 2015. Feeding preferences of oxpeckers in Kruger National Park, South Africa. Afr. Prot. Area Conserv. Sci. 57, 1–6.
|
|
Norval, R.A.I., Perry, B.D., Meltzer, M.I., Kruska, R.L., Booth, T.H., 1994. Factors affecting the distributions of the ticks Amblyomma hebraeum and A. variegatum in Zimbabwe: implications of reduced acaricide usage. Exp. Appl. Acarol. 18, 383–407.
|
|
Poulin, R., Mouillot, D., 2003. Parasite specialization from a phylogenetic perspective: a new index of host specificity. Parasitology 126, 473–480.
|
|
Robertson, A., Jarvis, A.M., 2000. Oxpeckers in north-eastern Namibia: recent population trends and the possible negative impacts of drought and fire. Biol. Conserv. 92, 241–247.
|
|
R Core Team, 2020. R: A language and environment for statistical computing. R
Foundation for Statistical Computing.
|
|
Stutterheim, I.M., Bezuidenhout, J.D., Elliott, E.G., 1988. Comparative feeding behaviour and food preferences of oxpeckers (Buphagus erythrorhynchus and B. africanus) in captivity. Onderstepoort J. Vet. Res. 55, 173–179.
|
|
Tarakini, T., Sithole, S., Utete, B., Muposhi, V., Madhlamoto, D., Gandiwa, E., 2017. Host preferences, spatial distribution and interaction of oxpeckers with wild ungulates in and around southern Gonarezhou National Park, Zimbabwe. Trop. Ecol. 58, 833–838.
|
|
Vamos, E.E., Elbrecht, V., Leese, F., 2017. Short COI markers for freshwater macroinvertebrate metabarcoding (No. e3037v2). PeerJ Preprints 5, e3037v2.
|
|
Ward, D., Robertson, A., 2017. Oxpeckers in Namibia: a review of their status and distribution in 2017. Namibian J. Environ. 1A, 6–13.
|
|
Weeks, P., 2000. Red-billed oxpeckers: vampires or tickbirds? Behav. Ecol. 11, 154–160.
|
| Phylum | Class | Order | Family | OTU | Oxpecker species | Localities | |||||||||
| Red-billed Oxpecker | Yellow-billed Oxpecker | Izumba Village | Ioma Village | Chobe River Lodge | |||||||||||
| Total=51 | Total=15 | Total=30 | Total=28 | Total=4 | |||||||||||
| N | % | N | % | N | % | N | % | N | % | ||||||
| Arthropoda | Arachnida | Araneae | Cheiracanthiidae | Cheiracanthium 1 | 0 | 0.00% | 1 | 6.67% | 0 | 0.00% | 0 | 0.00% | 1 | 25.00% | |
| Ixodida | Ixodidae | Amblyomma variegatum | 9 | 17.65% | 0 | 0.00% | 1 | 3.33% | 8 | 28.57% | 0 | 0.00% | |||
| Hyalomma truncatum | 1 | 1.96% | 1 | 6.67% | 1 | 3.33% | 0 | 0.00% | 0 | 0.00% | |||||
| Ixodidae 1 | 17 | 33.33% | 1 | 6.67% | 4 | 13.33% | 13 | 46.43% | 1 | 25.00% | |||||
| Rhipicephalus decoloratus | 23 | 45.10% | 5 | 33.33% | 6 | 20.00% | 19 | 67.86% | 3 | 75.00% | |||||
| Rhipicephalus evertsi | 11 | 21.57% | 4 | 26.67% | 5 | 16.67% | 8 | 28.57% | 2 | 50.00% | |||||
| Rhipicephalus zambeziensis | 2 | 3.92% | 0 | 0.00% | 1 | 3.33% | 1 | 3.57% | 0 | 0.00% | |||||
| Insecta | Coleontera | Tenebrionidae | Zophobas atratus | 1 | 1.96% | 1 | 6.67% | 2 | 6.67% | 0 | 0.00% | 0 | 0.00% | ||
| Diptera | Ceratopogonidae | Culicoides leucostictus | 1 | 1.96% | 0 | 0.00% | 0 | 0.00% | 1 | 3.57% | 0 | 0.00% | |||
| Not identified | Diptera 1 | 1 | 1.96% | 0 | 0.00% | 0 | 0.00% | 1 | 3.57% | 0 | 0.00% | ||||
| Diptera 2 | 1 | 1.96% | 0 | 0.00% | 1 | 3.33% | 0 | 0.00% | 0 | 0.00% | |||||
| Diptera 3 | 1 | 1.96% | 0 | 0.00% | 0 | 0.00% | 1 | 3.57% | 0 | 0.00% | |||||
| Diptera 4 | 16 | 31.37% | 9 | 60.00% | 19 | 63.33% | 2 | 7.14% | 1 | 25.00% | |||||
| Chordata | Mammalia | Artiodactyla | Bovidae | Aepyceros melampus | 2 | 3.92% | 0 | 0.00% | 0 | 0.00% | 2 | 7.14% | 0 | 0.00% | |
| Bos taurus | 47 | 92.16% | 14 | 93.33% | 25 | 83.33% | 28 | 100.00% | 4 | 100.00% | |||||
| Hippotragus niger | 2 | 3.92% | 0 | 0.00% | 2 | 6.67% | 0 | 0.00% | 0 | 0.00% | |||||
| Syncerus caffer | 2 | 3.92% | 2 | 13.33% | 3 | 10.00% | 0 | 0.00% | 1 | 25.00% | |||||
| Giraffidae | Giraffa camelopardalis | 7 | 13.73% | 0 | 0.00% | 2 | 6.67% | 5 | 17.86% | 0 | 0.00% | ||||
| Perissodactyla | Equidae | Equus burchellii | 6 | 11.76% | 0 | 0.00% | 6 | 20.00% | 0 | 0.00% | 0 | 0.00% | |||