| Citation: | Kai Song, Tom van der Valk, Bin Gao, Peter Halvarsson, Yun Fang, Wendong Xie, Siegfried Klaus, Zhiming Han, Yue-Hua Sun, Jacob Höglund. 2024: Inbreeding and genetic load in a pair of sibling grouse species: Tetrastes sewersowi and T. bonasia. Avian Research, 15(1): 100184. DOI: 10.1016/j.avrs.2024.100184 |
Genetic load and inbreeding are recognized as important factors to be considered in conservation programs. Elevated levels of both can increase the risk of population extinction by negatively impacting fitness-related characters in many species of plants and animals, including humans (inbreeding depression). Genomic techniques are increasingly used in measuring and understanding genetic load and inbreeding and their importance in evolution and conservation. We used whole genome resequencing data from two sibling grouse species in subarctic Eurasia to quantify both. We found a large range of inbreeding measured as FROH (fraction of runs of homozygosity) in individuals from different populations of Chinese Grouse (Tetrastes sewerzowi) and Hazel Grouse (T. bonasia). FROH estimated from genome-wide runs of homozygosity (ROH) ranged from 0.02 to 0.24 among Chinese Grouse populations and from 0.01 to 0.44 in Hazel Grouse. Individuals from a population of Chinese Grouse residing in the Qilian mountains and from the European populations of Hazel Grouse (including samples from Sweden, Germany and Northeast Poland) were the most inbred (FROH ranged from 0.10 to 0.23 and 0.11 to 0.44, respectively). These levels are comparable to other highly inbred populations of birds. Hazel Grouse from northern China and Chinese Grouse residing in the Qinghai-Tibetan Plateau showed relatively lower inbreeding levels. Comparisons of the ratio between deleterious missense mutations and synonymous mutations revealed higher levels in Chinese Grouse as compared to Hazel Grouse. These results are possibly explained by higher fixation rates, mutational melt down, in the range-restricted Chinese Grouse compared to the wide-ranging Hazel Grouse. However, when we compared the relatively more severe class of loss-of-function mutations, Hazel Grouse had slightly higher levels than Chinese Grouse, a result which may indicate that purifying selection (purging) has been more efficient in Chinese Grouse on this class of mutations.
Inbreeding (mating between relatives) is a major concern for conservation as it may lead to inbreeding depression (Wright, 1977; Shields, 1987) and fixation of harmful mutations (Lynch and Gabriel, 1990), thereby decreasing individual fitness which will increase the risk of population extinction (Soulé, 1980; Frankham, 1995a,b; Newman and Pilson, 1997; Charlesworth and Willis, 2009; Kardos et al., 2018). Inbreeding may become especially prevalent over time in small, fragmented and isolated populations (Höglund, 2009). Therefore, habitat fragmentation is likely to increase extinction proneness (Frankham, 1995a,b; Frankham, 1995a,b). The manifestation of inbreeding depression is determined by the segregating mutation load in any given population (van Oosterhout, 2020; Bertorelle et al., 2022). Since mutations are more common in large populations, by the fact that they contain more individuals, mutation load is expected to be positively correlated with population size (Kardos et al., 2021). In large populations, however, the effects of deleterious mutations will be partly or wholly phenotypically masked by favorable genetic variants in heterozygote diploid organisms (Kardos et al., 2021; Kyriazis et al., 2021). Thus, in large populations the "segregation load" increases. When populations become small and inbred, such mutations may become unmasked, causing inbreeding depression and purifying selection may purge them from the population (Bertorelle et al., 2022; Mathur et al., 2022; Smeds and Ellegren, 2022). A process which is affected by the severity of the fitness effect of the mutation, i.e. the selection intensity, is stronger on serious mutations (Kyriazis et al., 2021). However, some deleterious mutations may become fixed by genetic drift (sometimes referred to as mutational meltdown) and hence the "fixation load" may increase in small populations (van Oosterhout, 2020; Bertorelle et al., 2022). What load to expect in populations of conservation concern is thus a complex interplay between the demographic history such as fragmentation and population size fluctuations and the strength of purifying selection (Hedrick and Garcia-Dorado, 2016; Kardos et al., 2021).
The Chinese Grouse (Tetrastes sewerzowi) and Hazel Grouse (T. bonasia) are a pair of sibling species of the genus Tetrastes inhabiting boreal forests in the Qinghai-Tibetan Plateau (QTP) in the case of Chinese Grouse (Sun, 2000) and the Eurasian taiga in the case of Hazel Grouse (Storch, 2000). The Chinese Grouse has a narrow and fragmented distribution in the southeastern edge of the QTP, as well as a small long-term isolated population in the Qilian Mountains (Sun, 2000; Sun et al., 2003; Song et al., 2020). The Hazel Grouse is distributed in the large boreal forest region in northern and central Eurasia (Vaurie, 1965; Storch, 2000). The Hazel Grouse is classified as Lower Risk (Least Concern) overall by the International Union for the Conservation of Nature (IUCN) (IUCN, 2020), but it is on the red-list in some central and southern European countries because local populations have become extinct or are declining over large portions of its range in Europe (Swenson, 1991; Rózsa et al., 2016). The effective population size of Hazel Grouse in the Far East is large, but it is lower in the European populations which were bottlenecked during the Last Glacial Maximum (Song et al., 2021).
Here, we used whole genome resequencing data from individuals from different parts of the distribution range of both species to quantify inbreeding and genetic load. Importantly, both of these species have limited dispersal capabilities with narrow habitat requirements and some populations have been long-term isolated and have experienced bottlenecks (Storch, 2000). We inferred inbreeding levels by analyzing runs of homozygosity (ROH) for each individual. We then estimated genetic load for each individual by estimating the proportion of homozygous-derived mutations in regions of the genomes for deleterious missense mutations and the more severe class of loss-of-function mutations. Finally, we determined which genes have been fixed for both classes of mutations in Chinese Grouse. Our aim was to study how genetic load is shaped by the kind of mutations segregating in natural populations, as well as the demographic history and present threat status of the populations.
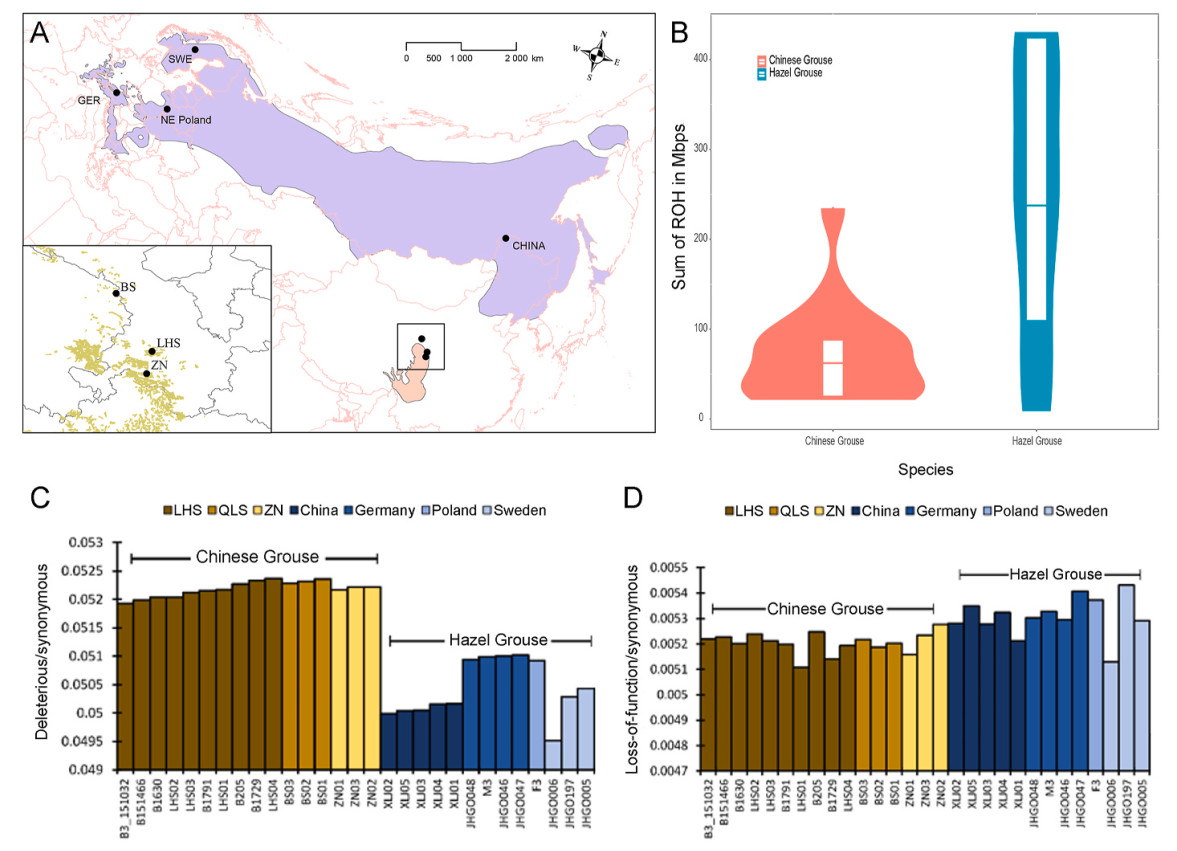
A total of 29 individual grouses from 8 locations were collected, 16 from three Chinese Grouse populations (3 from the Qilian Mountains (QLS); 3 from Zhuoni (ZN); 10 from the Lianhuashan National Nature Reserve (LHS)) along with 13 individuals from four Hazel Grouse populations (1 from Northeast Poland (NEP); 1 from the Austrian Alps and 3 from Bavarian forests in Germany (GER); 3 from Jämtland, Sweden (SWE); and 5 from Northeast China (XLJ), Fig. 1A, Appendix Table S1). Blood or muscle tissue was collected and preserved in 99% ethanol and stored at −20 ℃. Genomic DNA was extracted using DNeasy Blood & Tissue kit (Qiagen, Germany) (Song et al., 2021).
All samples were sequenced using the Illumina 150 PE sequencing strategy at Annoroad Gene Technology (Beijing, China). DNA libraries were constructed according to the manufacturer's recommendations.
All FASTQ data were adapter and quality trimmed using Trimmomatic with recommended settings (Bolger et al., 2014) and mapped against the chicken (Gallus gallus, GRCg6a) reference genome (Warren et al., 2017) using bwa-mem (Li, 2013) with default settings. We mapped to an outgroup species to ensure our results are not affected by reference bias arising from mapping to an ingroup reference. Although mapping to an outgroup reference may result in underrepresentation of fast evolving sites, genetic load estimates will not be biased toward any of the studied taxa, as gene order and synteny are generally conserved among birds (Backström et al., 2006; Hale et al., 2008). Chicken and the two grouse species studied here belong to the same avian family "Phasianidae" and have been separated for about 30 million years (Persons et al., 2016). Samtools was used to filter out reads below a mapping quality of 30 (phred-scale) (Li, 2011). Next, reads were realigned around indels using GATK IndelRealigner (McKenna et al., 2010) and we marked duplicates using Picard2.10.3 (https://broadinstitute.github.io/picard/). Next, we called single nucleotide polymorphisms (SNPs) with GATK UnifiedCaller outputting all sites (Danecek et al., 2011). Raw variant calls were then hard filtered following the GATK best practices (Van der Auwera et al., 2013). Additionally, we removed all SNPs below quality 30 (phred-scale), those with more than three times average genome-wide coverage across the data set. Sites for which > 75% of samples had a non-reference allele in a heterozygous state, indels, and sites within highly repetitive regions were identified from the repeatmask-track for the GRCg6a reference using VCFtools (Danecek et al., 2011).
To evaluate the level of inbreeding, runs of homozygosity (ROH) and the fraction of ROHs (FROH) were called using BCFtools/RoH (Narasimhan et al., 2016), a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. We restricted this analysis to scaffolds identified during the mapping stage by using SAMtools view on each mapped file (Li et al., 2009). Variant call files (VCF) were generated using bcftools (Danecek et al., 2011) mpileup with flags "-Ou" and subsequently indels were filtered out with flags-remove-indels. Indels were skipped during this step, because genotype calls in these regions tend to be enriched for errors, due to low mapping quality and mismapping. We filtered these files for sites with a depth greater than of 10 × and with a quality score over 30 and –maf 0.05 in the population level. Subsequently, we ran BCFtools RoH with flags "-G 30" and "—AF-dflt 0.4" to specify the use of genotype calls with a quality of 30 or more and to set a default allele frequency. Because of small per population sample size, we decided to fix the alternative allele frequency (option —AF-dflt) to 0.4.
To estimate genetic load in the Chinese and Hazel Grouse, we used the mappings and genome annotation of the chicken (GRCg6a). The variant effect predictor tool (McLaren et al., 2016) was used to identify loss-of-function mutations (transcript ablation, splice donor variant, splice acceptor variant, stop gained, frameshift variant, inframe insertion, inframe deletion, and splice region variant), missense, and synonymous mutations on the filtered SNP calls. SIFT was then used to identify those missense mutations which are likely deleterious (Ng and Henikoff, 2003). As an indication of mutational load, for each individual, we counted the number of genes containing one or more loss-of-function and the total number of deleterious missense mutations and divided both by the number of synonymous mutations (dN/dS) (Fay et al., 2001). We excluded all missense mutations within genes containing a loss-of-function mutation, as these are expected to behave neutrally. Dividing by the number of synonymous mutations mitigates species-specific bias, such as mapping bias because the reference genome was derived from the chicken, coverage differences, and mutation rate.
We identified all missense mutations where all Chinese Grouses carry the derived allele in the homozygous state and all Hazel Grouses with the ancestral allele or vice versa (using the chicken-reference as proxy for the ancestral state). Genes were then ranked by the number of fixed derived alleles they carry in each of the species.
To explore the evolution of functional categories, we used Kobas (Xie et al., 2011) to annotate the genes under selection in each of the species using the chicken reference genome (GRCg6a). These genes were submitted to Gene Ontology and KEGG databases for enrichment analysis. We used a false discovery rate (FDR) corrected binomial distribution probability approach to test significant enrichment gene function at a level of P < 0.05 (Benjamini and Hochberg, 1995).
There was a large range of FROH in both species. The range was from 0.02 to 0.24 among Chinese Grouse individuals and from 0.01 to 0.43 among Hazel Grouse (Appendix Table S1). This demonstrates that some populations of both Chinese Grouse and Hazel Grouse populations displayed relatively high levels of inbreeding. This was especially so for individuals from the QLS population of Chinese Grouse and European populations of Hazel Grouse (SWE, GER and NEP). Individuals from the other populations of Chinese Grouse (LHS and ZN) and Hazel Grouse (XLJ) displayed lower inbreeding levels (Appendix Fig. S1, Table S1). On average, Chinese Grouse showed lower inbreeding than Hazel Grouse (Student's t = 3.01, df = 27, P < 0.006, Fig. 1B).
The ratio of deleterious missense mutations to synonymous mutations was more prevalent in the range-restricted Chinese Grouse (Student's t = 171.0, df = 27, P < 0.001, Fig. 1C) as compared to Hazel Grouse. However, when we compared the ratio between loss-of-function and synonymous mutations, Hazel Grouse had slightly higher genetic load (Student's t = 4.55, df = 27, P < 0.001, Fig. 1D) but the ratio of this class of mutations was an order of magnitude less than that of deleterious missense mutations (see also Appendix Table S2).
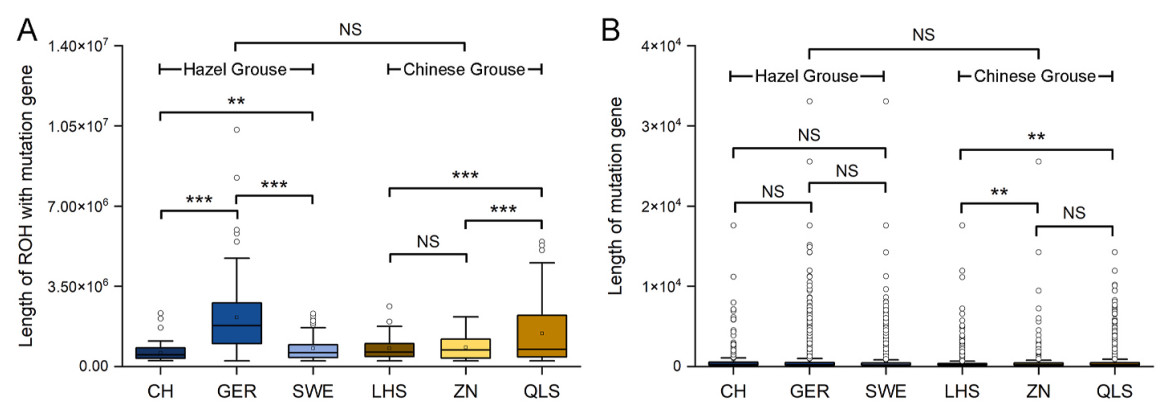
We inferred the relative length and abundance of ROHs with mutations and the positions of mutations for each individual on the longest scaffold of a Chinese Grouse assembly (Song et al., 2020) and found slightly longer such ROHs in Hazel Grouse than in Chinese Grouse (Nested ANOVA, F = 12.04, df = 1, P < 0.001, Fig. 2A). In Hazel Grouse, we found that mutations were evenly spread both inside and outside ROHs (e.g., contig 20 harbor many mutations, Appendix Fig. S1) and longer ROHs in the Swedish population (Nested ANOVA, F = 30.3, df = 2, P < 0.001, Fig. 2A). In Chinese Grouse, we found some ROHs absent of mutations (e.g., contig 20, 24, 27, 28 and 31, Appendix Fig. S1) and shorter ROHs in the relatively larger and more connected ZN population (Nested ANOVA, F = 31.62, df = 2, P < 0.001, Fig. 2A). The length of the genes carrying mutations were on average slightly longer in Hazel Grouse compared to Chinese Grouse (Nested ANOVA, F = 234.4, df = 1, P < 0.001, Fig. 2B). These results indicate that ROHs containing mutations have been purged from the genomes of Chinese Grouse.
We identified the genes fixed in Chinese Grouse which carried missense mutations and found that these included genes linked to disease resistance by being identified to be associated to anemia, cytokine receptors, influenza and herpes virus as well as homologous recombination (Appendix Tables S3 and S4).
In this study, we compared inbreeding levels and genetic load in Hazel Grouse that has a large and wide-ranging distribution and Chinese Grouse that has a more narrow and restricted distribution. If range distribution scales with population size one would thus expect both census and effective population size to have been lower through long periods of time in Chinese Grouse compared to Hazel Grouse, a prediction not entirely borne out in a recent study of the demographic history of the two species (Song et al., 2021). Instead, PSMC analyses (Li and Durbin, 2011) using genome-wide data from the same 29 individuals from these sibling Tetrastes species as in this study revealed complex demographic scenarios, with each species showing different rises and declines in effective population size (Song et al., 2021). Importantly, MSMC analyses (Schiffels and Durbin, 2014) indicated recent declines in populations of both species. Here, we estimated ROHs and genetic load for each individual to quantify realized genomic inbreeding and to identify genomic regions that may contribute to inbreeding depression and which may have been subjected to purging.
In general, the levels of ROH among populations is expected to be related to conservation concern with smaller and fragmented populations having more and longer ROHs and large and connected populations fewer and shorter ROHs (Ceballos et al., 2018). Our results suggest that Chinese Grouses on average are not more inbred than Hazel Grouse but with larger individual variation. Within species, European populations of Hazel Grouse were significantly more inbred when comparing ROHs in other populations. The effective population size of European populations of Hazel Grouse was lowered repeatedly during the last ice age 20 Kya which led to fragmented and isolated populations, which had to recolonize Northern Europe (Sweden and Germany) (Ingolfsson et al., 2006; Song et al., 2021). In contrast, the XJL population of Hazel Grouse resided in an area which was a refugium during the last ice age, resulting in a relatively large outbred population at present (Abbott et al., 2000; Saarnisto and Lunkka, 2004). In Chinese Grouse, the QLS population has experienced persistent low effective population sizes in a small isolated population at the northeast edge of the QTP, the Qilian Mountains (Zhan et al., 2011; Song et al., 2021). We found a larger fraction of the genomes in ROH in this population which suggests the QLS population has suffered inbreeding for a long time but, despite this, has survived for many generations at small effective population size (Keller and Waller, 2002; Charlesworth and Willis, 2009).
Purging can reduce genetic load and the depression of fitness through more efficient purifying selection facilitated by inbreeding (Hedrick, 1994; Tallmon et al., 2004; García-Dorado, 2012). Identifying the efficacy of purifying selection in large and small populations of endangered species could therefore aid in animal conservation (van der Valk et al., 2019a,b). When we estimated the ratio between deleterious missense and synonymous mutations (the more benign class of mutations studied here), Chinese Grouse displayed higher load than Hazel Grouse, possibly indicating mutation accumulation (mutational melt down) (Lynch and Gabriel, 1990) in the range-restricted species. However, when we measured the ratio between loss-of-function and synonymous mutations (the more severe class of mutations), purifying selection seemed to have been operating since this class of mutations were an order of magnitude less common than deleterious missense mutations. However, there was no large difference for this class of mutations when comparing Chinese Grouse and Hazel Grouse although the range-restricted Chinese Grouse displayed slightly less load. Deleterious missense mutations should be less deleterious compared to loss-of-function mutations (van der Valk, 2019) which should imply stronger selection intensities (purging) on the latter. The KEGG-pathway and Gene Ontology analyses further suggest that the mutations that have become fixed in Chinese Grouse may be important for the organism and may even have been subjected also to positive selection as our analyses identified that many of them were found in genes associated with anemia, cytokine receptors, influenza and herpes virus as well as homologous recombination in model organisms.
Our population genomic analyses thus indicated that the range-limited Chinese Grouse had a lower mutational load when loss-of-function mutations are considered than the wider ranging Hazel Grouse. A result in line what has been found when comparing large and small populations in other studies such as those of Mountain Gorillas (Gorilla beringei beringei) (Xue et al., 2015), Channel Island Fox (Urocyon littoralis) (Robinson et al., 2018), Alpine Ibex (Capra ibex) (Grossen et al., 2020), the Kākāpō (Strigops habroptilus) (Dussex et al., 2021), Indian tigers (Khan et al., 2021), European and Iberian Lynx (Lynx pardinus) and Ethiopian Wolf (Canis simensis) (Mooney et al., 2023). These patterns have generally been explained by more efficient purging in smaller populations, a scenario which may also apply for loss-of-function mutations in the case of the two sibling species of grouse in our study. The fact that some ROHs in Chinese Grouse are completely devoid of mutations speaks in favor of such an interpretation. However, contrary to the studies cited above, which found the same pattern of reduced load also for missense mutations, we found that deleterious missense mutation load was higher in Chinese Grouse than in Hazel Grouse. This indicates that missense load may accumulate more easily (Feng et al., 2019; Hedrick et al., 2019; van der Valk et al., 2019a,van der Valk et al., 2019b; Szpiech et al., 2013) whereas the more serious fitness effects imposed by loss-of-function mutations may become removed by purging (Wootton et al., 2023). Our results thus indicate that genetic load is shaped by a complex interplay of the kind of mutations segregating in natural populations and their demographic history and present threat status.
This study was approved by the Animal Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences (IOZ20150069). The blood samples were collected from wild Chinese Grouse, which were released back in to the wild. The procedure of blood collection was in strict accordance with the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
Sequencing data for the Chinese Grouse and Hazel Grouse have been deposited in Short Read Archive under project number PRJNA588719 and PRJCA005913.
Kai Song: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Tom van der Valk: Writing – original draft, Methodology, Formal analysis. Bin Gao: Writing – original draft, Methodology, Formal analysis. Peter Halvarsson: Writing – original draft, Supervision, Formal analysis, Conceptualization. Yun Fang: Writing – review & editing, Data curation, Conceptualization. Wendong Xie: Writing – original draft, Formal analysis. Siegfried Klaus: Writing – review & editing, Data curation, Conceptualization. Zhiming Han: Writing – review & editing, Supervision, Conceptualization. Yue-Hua Sun: Writing – review & editing, Supervision, Methodology, Funding acquisition, Conceptualization. Jacob Höglund: Writing – review & editing, Writing – original draft, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
We would like to thank the Annoroad Gene Technology in Beijing for performing the whole genome sequencing, and Ying-Xin Jiang for his help in sample collection.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.avrs.2024.100184.
|
Höglund, J., 2009. Evolutionary Conservation Genetics. Oxford University Press, Oxford.
|
|
Ingolfsson, O., Moller, P., Lubinski, D., Forman, S., 2006. Severnaya Zemlya, Arctic Russia: a Nucleation Area for Kara Sea Ice Sheets During the Middle to Late Quaternary. AGU Fall Meeting Abstracts.
|
|
Kardos, M., Åkesson, M., Fountain, T., Flagstad, Ø., Liberg, O., Olason, P., et al., 2018. Genomic consequences of intensive inbreeding in an isolated wolf population. Nat. Ecol. Evol. 2, 124-131.
|
|
Li, H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997.
|
|
Mathur, S., Tomeček, J.M., Tarango-Arámbula, L.A., Perez, R.M., DeWoody, J.A., 2022. An evolutionary perspective on genetic load in small, isolated populations as informed by whole genome resequencing and forward-time simulations. Evolution 77, 690-704.
|
|
Saarnisto, M., Lunkka, J.P., 2004. Climate variability during the last interglacial-glacial cycle in NW Eurasia. In: Battarbee, R.W., Gasse, F., Stickley, C.E. (Eds.), Past Climate Variability through Europe and Africa, vol. 6. Springer, Dordrecht, pp. 443–464.
|
|
Shields, W.M., 1987. Dispersal and mating systems: investigating their causal connections. In: Chepko-Sad, B.D., Halpin, Z.T. (Eds.), Mammalian Dispersal Patterns: the Effects of Social Structure on Population Genetics. University of Chicago Press, Chicago, pp. 3–24.
|
|
Smeds, L., Ellegren, H., 2022. From high masked to high realized genetic load in inbred Scandinavian wolves. Mol. Ecol. 32, 1567-1580.
|
|
Soulé, M.E., 1980. Conservation Biology: An Evolutionary Ecological Perspective. Sinauer Associates Inc., Sunderland, MA.
|
|
Van der Auwera, G.A., Carneiro, M.O., Hartl, C., Poplin, R., Del Angel, G., LevyMoonshine, A., et al., 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 43 (11), 10.11-11.10.33.
|
|
van der Valk, T., 2019. Genomics of Population Decline. Doctoral Dissertation. Uppsala Unversity, Uppsala.
|
|
Vaurie, C., 1965. The birds of the Palearctic fauna. Non-passeriformes. Bird-Banding 37, 145-146.
|
|
Wright, S., 1977. Evolution and the Genetics of Populations. 2. The Theory of Gene Frequencies. University of Chicago Press, Chicago, IL.
|