| Citation: | Zhehan Dong, Shangmingyu Zhang, Yuwen Cheng, Xingcheng He, Ian Haase, Yi Liang, Yong Jiang, Yongjie Wu. 2022: Comparative analysis of the intestinal tract microbiota and feeding habits of five sympatric flycatchers. Avian Research, 13(1): 100050. DOI: 10.1016/j.avrs.2022.100050 |
Gut microbiota and host interactions co-evolve and develop into stably adapted microbial communities and play vital roles in maintaining the health of organisms. Diet is supposed to be an important driver of differences in gut microbiota, but previous studies would commonly use literature depictions, which are essential but inaccurate, to explain the effects of diet on the gut microbiota of wild birds. In this study, we collected intestinal samples from five sympatric flycatchers to compare the gut microbial differences using bacterial 16S rRNA genes from Illumina MiSeq platform. Over 1,642,482 quality-filtered sequences from 18 16S rRNA libraries were obtained and distinct compositions and diversities of gut microbiota were found in five flycatchers. Their gut microbiota is mainly from the four bacterial phyla of Proteobacteria, Firmicutes, Actinomycetes, and Bacteroidetes, but at the genus level showed a significant difference. Functional predictions revealed that the metabolic capacity of the gut microbiota of five flycatchers is greatly distinguished at KEGG level 3. And multiple food fragments showed a significant correlation with gut microbiota. Besides, the significant differences in the specific composition of the diets of the five insectivorous flycatchers indicated the differentiation of dietary niches. The study of the gut microbiota and feeding habits of sympatric flycatchers would increase the understanding of the gut microbial diversity of wild birds, and also improve our cognition of the co-evolution and co-adaptation within the host gut microbiota relations.
Microbiota is the collection of microorganisms that live in a particular environment. Diverse microbial communities are an important part of life and contribute to the evolution of life (Burge, 1988; Ley et al., 2008). Animals have never been autonomous organisms, but rather co-existed and evolved with many different species of microbes in their body (Bordenstein and Theis, 2015). The association between an animal and its gut microbiota is a complex symbiosis and the gut microbiota influences host health, physiology, development, and adaptation (Ley et al., 2008; Al-Asmakh et al., 2014; Capunitan et al., 2020a). The gut microbiota of animals begin to form shortly after birth and have been shown to be remarkably host-specific and remain generally stable over time (Faith et al., 2013). However, the colonization of these microbial communities can be influenced by both the host immune system and gut physiology (Kelly et al., 2007). Throughout the host's life, ecological factors (environmental filtering) such as diet, habitat, and social interactions further affect gut microbiota composition and stability (i.e., individual variation and turnover rates) of gut microbiota. Thus, to understand the evolution and the long-term associations between hosts and their gut microbes, we need to explore the relative importance of these ecological factors in affecting gut microbial communities.
Among multiple ecological factors, diet appears to have a strong influence on shaping the gut microbiota of many animals (Muegge et al., 2011; Pan and Yu, 2014; Youngblut et al., 2019), especially in birds (Bodawatta et al., 2021a, b). Most of the studies on avian gut microbiota assigned bird taxa to particular dietary guilds based on literature (Wilman et al., 2014; Hird et al., 2015; Bodawatta et al., 2021; Bodawatta et al., 2022; Hou et al., 2021). However, these studies ignored individual and seasonal natural variation in bird diet (Evans et al., 1997) and the variation of diet may be an important factor for the gut microbiota composition (Góngora et al., 2021). Besides, overall dietary guild assignments (e.g. frugivore and insectivore) tend to assort closely related species into a group which ignore the niche differentiation and diet difference among closely related species (Burin et al., 2016). Considering that host taxonomy and diet are two independent factors influencing gut microbiota and birds within the same feeding guild and closely related species often consume diverse combinations of foods, it is necessary to examine the natural diet composition of bird individuals and its influence on gut microbiota. However, such research is currently lacking in gut microbiota research of birds (Bodawatta et al., 2022b).
Birds are a highly suitable group to study host-microbiota associations because there are over 10,000 species and their diet and migration behavior are quite different in their life history. Previous studies on gut microbiota have mainly focused on humans and mammals, while the studies on birds only account for 10% of all studies on gut microbiota and which were mainly focused on poultry or zoo birds (Grond et al., 2018). The use of captive animals allows for controlled experiments that have led to significant advances in our understanding of their biology. However, due to the captive environments differ from natural and wild environments in diet, habitat and exposure to different environmental microbes. Previous study demonstrated that the microbiotas of captive birds are likely different from those of wild birds (Amato et al., 2019). Therefore, conducting studies on wild species is essential to understand the importance of multiple factors in mediating gut microbiota diversity in natural ecological settings among avian species.
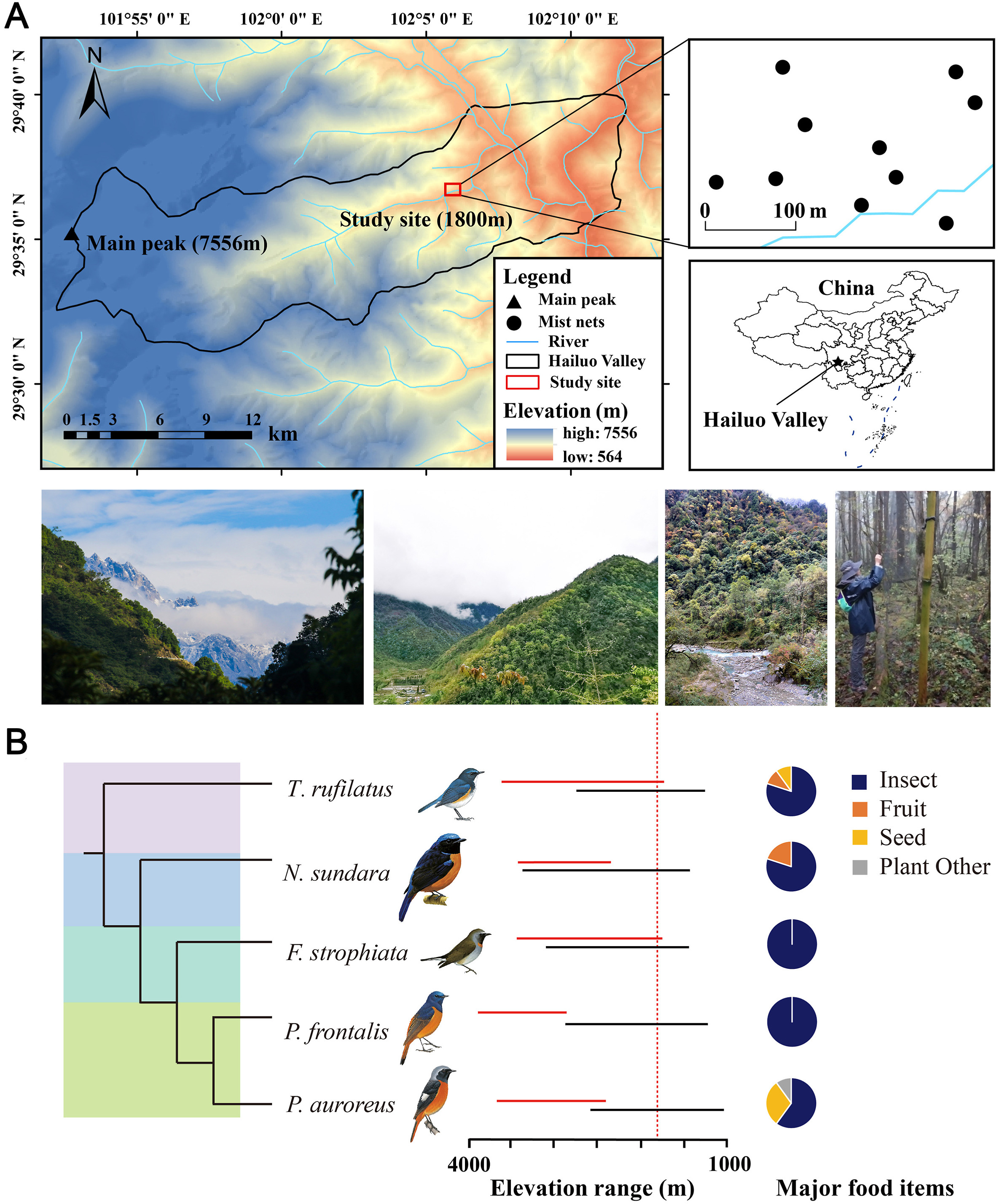
The Old World Flycatchers are the largest family of passerine birds, which are abundant and widely distributed, and play an important role in forest ecosystems (Sangster et al., 2010). Their foraging ecology contributes to the regulating and supporting ecosystem services, including nutrient cycling, seed dispersal, and pest control (Whelan et al., 2015). Rufous-gorgeted Flycatcher (Ficedula strophiata), Daurian Redstart (Phoenicurus auroreus), Rufous-bellied Niltava (Niltava sundara), Blue-fronted Redstart (Phoenicurus frontalis), and Himalayan Bluetail (Tarsiger rufilatus) are the five most abundant species among the 42 species of Muscicapidae distributed on the eastern slopes of Mount (Mt.) Gongga (Wu et al., 2017). They belong to four different genera and are closely related in Muscicapidae, with sexually dichromatic and socially monogamous (Morimoto et al., 2006). All five species engage in altitude migration and migrate from non-breeding areas to breeding areas at different elevations which is a less studied but widespread behavior in montane animals (Barçante et al., 2017; Hsiung et al., 2018). The five species are distributed in similar elevation ranges both during the breeding and non-breeding seasons (Fig. 1B). They are supposed to be insectivores and as they have typical insectivorous digestive tract morphology, including developed proventriculus, ventriculus with thick gastric cuticula, and a short cecum (Richardson & Wooller, 1986). In addition to small invertebrates, their diets also include some plants-based food, such as fruits, seeds, and other parts of a plant (Fig. 1B).
The study of the coexistence mechanism of sympatric species has always been a fascinating subject of community ecology. According to the competitive exclusion principle, two complete competitor species cannot coexist in the same area at the same time under a limited resource (Gause, 1934). In practice, the five flycatchers have highly ecological overlap in Mt. Gongga. However, competition can be increased between sympatric congeners due to their similarities, particularly. To avoid the ecological competition, closely related species commonly inhabit different habitats or microhabitats and these differences be observed significantly in their diet composition (Pianka, 2011). Therefore, the investigation of diet composition is one of the first steps to better understanding the ecological mechanisms involved in the avoidance of competition between closely related sympatric species.
Hereby we collected tissue samples from five sympatric insectivorous flycatchers in Hailuo Valley of Mt. Gongga. By analyzing the gut bacterial 16S rRNA genes from MiSeq amplicon sequencing and microscopy of the stomach content, several goals are expected to be achieved including: (1) to analyze the gut microbial communities and explore the metabolism and function of gut microbiota of the five flycatchers, (2) to investigate the diet divergence of sympatric birds in the same region, and (3) to explore the relationship between food composition and gut microbes. This study focuses on ecologically important but often overlooked taxa to increase our understanding of gut microbial diversity within and among bird species. This understanding would help us reveal how various host and environmental factors interact to shape the gut microbiota.
All animal research procedures strictly complied with the P.R. China Legislation on the Use and Care of Laboratory Animals and were approved by the Animal Care Review Committee, College of Life Sciences, Sichuan University, China.
Mt. Gongga (7556 m) is the highest mountain in the Hengduan Mountains. It is one of the top 36 biodiversity hotspots in the world and is located in the transitional zone between the Sichuan Basin and the Qinghai-Tibetan Plateau (Hrdina and Romportl, 2017). Hailuo Valley is located on the eastern slope of Mt. Gongga and has an altitude difference of 6400 m within a horizontal distance of 30 km (Zu et al., 2019). The complex topography and diverse habitat types harbor rich plant and insect diversity which provides a high diversity of food sources for the rich small passerine birds.
Five different wild bird species were collected in Hailuo Valley on the eastern slope of Mt. Gongga in Sichuan Province (Fig. 1A) from April to May 2021: (i) Rufous-gorgeted Flycatcher (Ficedula strophiata), (ii) Daurian Redstart (Phoenicurus auroreus), (iii) Rufous-bellied Niltava (Niltava sundara), (iv) Blue-fronted Redstart (Phoenicurus frontalis) and (v) Himalayan Bluetail (Tarsiger rufilatus). The phylogenetic relationship and ecological background of the five species of flycatchers, including distribution altitude, feeding habits, etc. (Fig. 1B), were obtained from the BirdTree (Jetz et al., 2012), the Birds of the World website (https://birdsoftheworld.org), EltonTraits 1.0 (Wilman et al., 2014) and historical bird survey data of Mt. Gongga of our research group. Birds were captured at the elevation of 1800 m using mist nets. Ten mist nets were set up at a distance of about 100 m from each other in an experimental sample plot of less than 0.06 km2 of the deciduous and evergreen broadleaved forest. Once captured birds of target species were immediately taken to the lab and euthanized. Carcasses were disinfected with 75% ethanol and opened ventrally from chest to abdomen to expose the entire gastrointestinal tract. The rectums of these birds are too short to store feces. We isolated the intestinal contents of the 6–7 cm jejunal-ileum segment above the rectum by sterile instruments, and transfer the contents into the cryovial for subsequent studies. The isolated intestine was immediately stored in liquid nitrogen and then transported back to the laboratory to separate the intestinal contents in the purifier vertical clean bench. The intestinal contents were extruded from one end of the intestine using sterile forceps, placed into cryovials, and stored in liquid nitrogen before DNA extraction. The stomach contents were separated from the bird's stomach, and the contents and stomach were stored separately in liquid nitrogen. The details of the samples used in the experiment are shown in (Table 1).
| Common name | Scientific name | Intestinal tract Reps | Intestinal tract Reps ID | Stomach contents Reps | Stomach contents Reps ID |
| Rufous-gorgeted Flycatcher | Ficedula strophiata | 3 | RF223, RF224, RF242 | 6 | RF195, RF223, RF224, RF227, RF242, RF256 |
| Daurian Redstart | Phoenicurus auroreus | 5 | DR63, DR65, DR87, DR91, DR122 | 6 | DR63, DR65, DR87, DR116, DR91, DR122 |
| Rufous-bellied Niltava | Niltava sundara | 4 | RN197, RN207, RN253, RN261 | 2 | RN207, RN253 |
| Blue-fronted Redstart | Phoenicurus frontalis | 3 | BR24, BR32, BR33 | 8 | BR18, BR24, BR27, BR28, BR32, BR36, BR94, BR98 |
| Himalayan Bluetail | Tarsiger rufilatus | 3 | HB45, HB92, HB96 | 5 | HB30, HB45, HB76, HB92, HB96 |
| Intestinal tract Reps were the number of intestinal tract sample replicates, and intestinal tract Reps ID corresponds to the individual number of the bird. Stomach contents Reps were the number of stomach contents sample replicates, and stomach contents Reps ID corresponds to the individual number of the bird. | |||||
Total genome DNA from samples was extracted using CTAB/SDS method. DNA concentration and purity was monitored on 1% agarose gels. According to the concentration, DNA was diluted to l μg/μL using sterile water. The highly variable V3–V4 region of the 16S rRNA gene was PCR-amplified from the extracted DNA using the primer pair 341F (5ʹ-CCTAYGGGRBGCASCAG-3ʹ) and 806R (5ʹ-GGACTACHVGGGTWTCTAAT-3ʹ) (Caporaso et al., 2012). The polymerase chain reaction (PCR) reactions were performed in a total of 50 μL volume, using 15 μL Phusion High-Fidelity PCR Master Mix with GC BufferAmpliTaq Gold DNA Polymerase (New England Biolabs Inc., Beijing, China), 3 μL primers and 5–10 ng of DNA template. The PCR conditions were 98 ℃ for 1 min, followed by 30 cycles of 10 s at 98 ℃; 30 s at 50 ℃ and 30 s at 72 ℃, and a final elongation step of 5 min at 72 ℃. The final amplicon pool was evaluated using the Fragment Analyzer (Advanced Analytics Technologies, Ames, USA). PCR products were also quantitatively assessed using a Qubit 2.0 Fluorometer (Life Technologies, USA), and diluted according to Illumina's standard protocol for Sequencing on Illumina MiSeq (Illumina Inc). The sequencing was performed by Health Time Gene Institute in Shenzhen, China.
The paired-end sequences of 16S rRNA genes were assembled using FLASH (v1.2.11) (Magoč and Salzberg, 2011). Read quality checking and filtering were done using the standard operating procedure with QIIME (v1.17) (Kuczynski et al., 2012). Amplicon sequencing chimeras were removed using UCHIME (v4.2.40) algorithm (Edgar et al., 2011). In VSEARCH (v2.4.4), we clustered the quality-filtered sequences into operational taxonomic units (OTUs) at a sequence similarity threshold of 97% (Rognes et al., 2016). OTUs were assigned and identified to taxa using the SLIVA (v132), GreenGene (v13.8) and Unite (v7) reference libraries and non-bacterial sequences (e.g. mitochondrial sequences) were removed (Quast et al., 2012).
We scraped and rinsed the entire contents from the stomachs, and left them to dry inside an oven at 50 ℃ for 24 h. Microscopic classification of undigested fragments was performed by Nikon SMZ745T stereo microscope. Since complete insect individuals have not been found in the stomach of birds, it is difficult to identify and classify the insects. Therefore, we used the method of fragment classification to compare stomach insect fragments (Hyslop, 1980). We sorted the same morphological structure or fragments with the same traits as the fragment classification unit (FCU). By comparing with the FCU library, the gastric contents of each individual were counted, and the relative abundance table of different fragments of gastric contents was obtained. The percentage of stomachs containing the same fragments (PAS) and relative abundance of these fragments (RA) were used to characterize the frequency of different fragment taxa in each bird species, which are defined as follows:
|
PAS=NiN0×100% |
(1) |
where Ni is the number of individuals that contain the same fragment in each species, and N0 is the number of individuals of each species.
|
RA=nin0×100% |
(2) |
where ni is the total number of a certain type fragment of each species, and n0 is the total number of fragments of each species.
The rarefaction curve was produced in alpha_rarefaction.py script in Qiime. Alpha-diversity analyses, including community richness parameters (Chao1 index) and community diversity parameters (Shannon-Wiener index) were calculated using Mothur. A Venn diagram was constructed based on these OTUs with the Venn diagram package (Chen & Boutros, 2011) in R (v3.1.1), showing the number of shared and unique OTUs among the different host species. Principal coordinate analysis (PCoA) was performed to summarize multidimensional clustering of microbial communities at the OTU level based on unweighted UniFrac distances and to evaluate the differences in food composition among five flycatchers based on Bray-Curtis distances. Linear discriminant analysis effect size (LEfSe) was conducted to identify genera differentially represented among five species. PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved states) v1.1.0 was used to detect predicted functional differences in gut microbiota between five flycatchers, and we used an ancestral state reconstruction algorithm to predict metagenomic functional profiles from 16S rRNA gene sequence data and a reference genome database. ANOVA and Wilcoxon rank-sum test were used to test for differences in on KEGG level 2 and 3, functional enrichment between species (Hyunjun Cho, 2019). Pearson's correlation analysis was used to examine the relationship between the proportion of insects in food and the relative abundance of Bacteroidetes in the gut. The Spearman's rho nonparametric correlations between the gut microbiota and FCUs were determined using psych packages. All statistical analyses were performed using the R statistical software package. Values smaller than p = 0.05 were statistically significant. Estimates of effect size allow the assessment of the strength of the relationship between the investigated variables and to determine the effect of one variable on another. Effect sizes used with parametric tests were calculated from ANOVA as eta squared (η2) (Tomczak and Tomczak, 2014), the coefficient can be calculated according to formula (3) according to the output result of aov function in R. In non-parametric tests, the eta squared (η2H) (Tomczak and Tomczak, 2014) statistic was computed from the Kruskal-Wallis test values. Effect sizes in correlation coefficient studies were calculated using Fisher's z transformed values (Borenstein et al., 2021).
The η2-statistic is calculated as:
|
η2=SSefSSt |
(3) |
where SSef is the sum of squares for the effect, and SSt is the total sum of squares.
The η2H-statistic is calculated as:
|
η2H=H−k+1n−k |
(4) |
where H is the value obtained in the Kruskal-Wallis test, k is the number of groups, and n is the total number of observations.
The z-statistic is calculated as:
|
z=0.5×ln(1+r1−r) |
(5) |
where r is the sample correlation coefficient, and z is the standardized sample correlation coefficient.
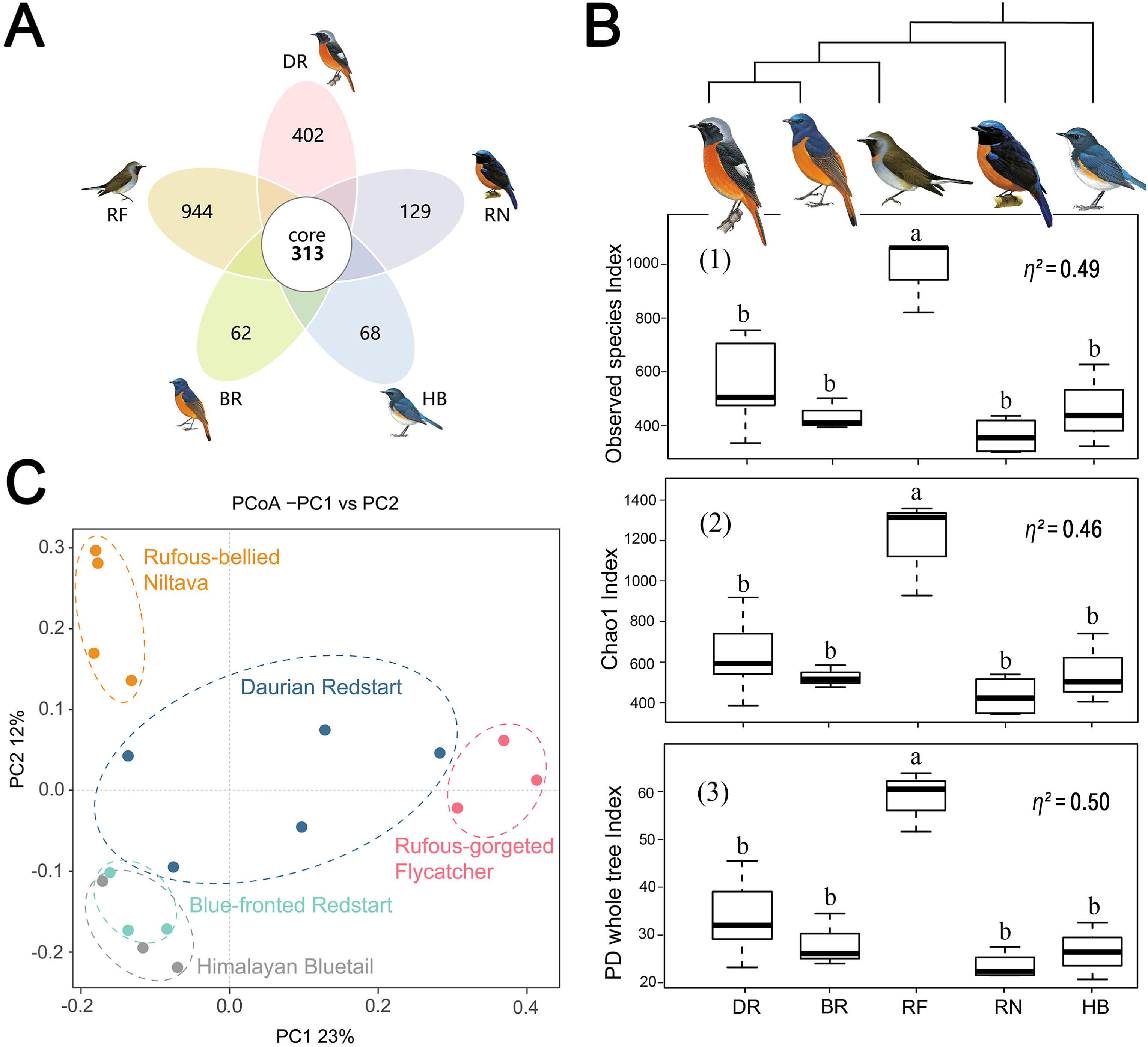
Across all samples, 1,642,482 sequences with an average length of 207 bp were obtained. The observed species rarefaction curve tended to plateau, which showed that the sequencing depth sufficiently captured the major microbiota of each sample (Appendix Fig. S1). In total, 3105 operational taxonomic units (OTUs) were identified through our quality control process and 313 OTUs were shared by all groups. The highest number of unique OTUs were found in the Rufous-gorgeted Flycatcher group (944 OTUs), followed by the Daurian Redstart group (402 OTUs). In contrast, the fewest unique OTUs were found in the Blue-fronted Redstart group (62 OTUs) and Himalayan Bluetail group (68 OTUs) (Fig. 2A).
We used observed species richness, Shannon index, Chao 1 index, and PD whole tree index to illustrate the alpha diversity of gut microbiota at the OTUs level. The α-diversity index (using observed species richness, Chao1 index, and PD whole tree index) of the Rufous-gorgeted Flycatcher group was the highest (t-test, P < 0.05), and significantly higher than the α-diversity index of the Daurian Redstart, Blue-fronted Redstart, Himalayan Bluetail, and Rufous-bellied groups. There was no significant difference among the other groups (Fig. 2B, Appendix Fig. S2).
PCoA showed that the gut microbiota are different among five species and it is more similar to each other within species than that among species (ANOSIM, R = 0.304, p = 0.01). More similarities were found between the Blue-fronted Redstart and the Himalayan Bluetail (Fig. 2C).
The predominant bacterial phyla present in the intestinal tracts of all birds were Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes (Fig. 3A). Bacteria from the phylum Firmicutes dominated the microbiota of the Rufous-gorgeted Flycatcher (61.82 ± 13.96%), while bacteria from the phylum Proteobacteria dominated the microbiota of the Himalayan Bluetail, Daurian Redstart, and Blue-fronted Redstart (58.61 ± 27.80%, 53.38 ± 21.63%, 52.87 ± 29.30%, respectively). Interestingly, compared to the Rufous-gorgeted Flycatcher, Fusobacteria were much less abundant in the other four flycatchers (2.36 ± 2.15% and < 0.03%, respectively). The same situation is true for Bacteroidetes, whose relative abundance was 0.18% in Blue-fronted Redstart and Himalayan Bluetail, while Bacteroidetes in the other three flycatchers were all higher than 5.62%.
At the genera level, the most dominant genera identified in the Daurian Redstart was Diplorickettsia (19.18%). Romboutsia and Clostridium.sensu.stricto.1 were the most dominant genera identified in Blue-fronted Redstart at a concentration of 12.65% and 6.72%, respectively. Escherichia and Enterococcus were the most dominant genera in Himalayan Bluetail, at a concentration of 27.67% and 13.33%, respectively. By contrast, the most dominant genera in Rufous-gorgeted Flycatcher and Rufous-bellied Nilltava were Faecalibacterium (3.83%), Serratia (3.63%) and Bacteroides (5.24%), Faecalibacterium (2.78%) (Fig. 3B). A large number of genera with relative abundance less than 1% were detected in each species, and these genera accounted for the total abundance 55.41–89.05% (Appendix Table S1).
The cladogram showed 6 phyla, 12 classes, 17 orders, 23 families, and 22 genera were dominant in distinct species (Fig. 4A). LEfSe analyses also showed that only in the Rufous-gorgeted Flycatcher, Fusobacteria were significantly more abundant compared to other species. The Daurian Redstart and Rufous-gorgeted Flycatcher groups showed the most unique microbiota characterized by the presence of diverse taxa. We found that 20 taxa were overrepresented in Daurian Redstart, e.g., Lactobacillales (order), Burkholderiales (order), Legionellales (order), and Bacteroidales (order). Twenty-seven taxa were overrepresented in Rufous-gorgeted Flycatcher, eg, Veillonellales (order), Fusobacteriales (order), Clostridiales (order), and Bacillales (order). The number of bacterial species that showed significant differences in the other three species was smaller. Enterococcaceae (family), Microbacterium (genus), Brucellaceae (family) showed higher abundance in Rufous-bellied Niltava, Himalayan Bluetail, Blue-fronted Redstart, respectively (Fig. 4B).
Predictive metagenomics analysis (PICRUSt) identified potential functional differences in the gut microbiota of five flycatchers. These differences include the low abundance of predicted gene content in KEGG pathways related to Cellular Processes and Organismal Systems, including Cell Communication and Sensory system, which were found in Blue-fronted Redstart and Himalayan Bluetail (Appendix Fig. S4). In addition to the above, five species exhibited similar gene functions at KEGG level 2, but more differences in functional enrichment of gut microbiota at KEGG level 3 (Appendix Table S2).
The unweighted Unifrac distance suggested that the bacterial community differences within each sample species were small, samples from the same species cluster together (with the exception of Daurian Redstart). Although the clustering relationship of the same species of microorganisms is relatively close, the similarity of the microbial clustering does not seem to have much connection with the phylogenetic key of birds (Appendix Fig. S5).
A total of 1747 undigested and identifiable fragments were found in the stomachs of 27 individuals through stereoscopic observation. After photographing all fragments and classifying the same fragments according to their shape characteristics, a total of 152 FCUs were separated. The five FCUs that appear most frequently in the stomach of different individuals of each species of bird were presented (Fig. 5). Although the five species of birds had some of the same food, they had different preferences for insect choices. Daurian Redstart has the highest diversity of food choices among the five species of birds, both in terms of observation value and Shannon index (Table 2). The PCoA plot shows that the five species of birds living in the same area showed some differences in food choices (ANOSIM, R = 0.26, p = 0.002). The stomach contents of Daurian Redstart, Rufous-gorgeted Flycatcher, Himalayan Bluetail, and Rufous-bellied Niltava are more closely clustered within the species. The diet of Blue-fronted Redstart did not show a strong clustering relationship among individuals (Appendix Fig. S6).
| Rufous-gorgeted Flycatcher | Daurian Redstart | Blue-fronted Redstart | Himalayan Bluetail | Rufous-bellied Niltava | |
| Observation | 41 | 69 | 38 | 33 | 18 |
| Shannon index | 2.68 | 3.03 | 2.91 | 0.90 | 2.47 |
| Observation was the number of FCUs with the same morphological characteristics and same traits that we actually counted in each species, Shannon index was used to demonstrate the diversity of the stomach contents, the higher value means the richer stomach contents. | |||||
The composition micro histological analysis revealed that the diet for Daurian Redstart, Rufous-gorgeted Flycatcher, and Rufous-bellied Niltava are almost insects, whereas the diet for Blue-fronted Redstart and Himalayan Bluetail was a mixture of insects (> 70%) and plant fruits (< 30%). To further investigate whether the gut microbiota composition is associated with the food composition, we conducted a correlation analysis between the proportion of insects in food and the relative abundance of Bacteroidetes in the gut (Appendix Fig. S7). The results showed that the relative abundance of Bacteroidetes in these birds was significantly correlated with the ratio of insects (r = 0.53, p = 0.036, z = 0.59).
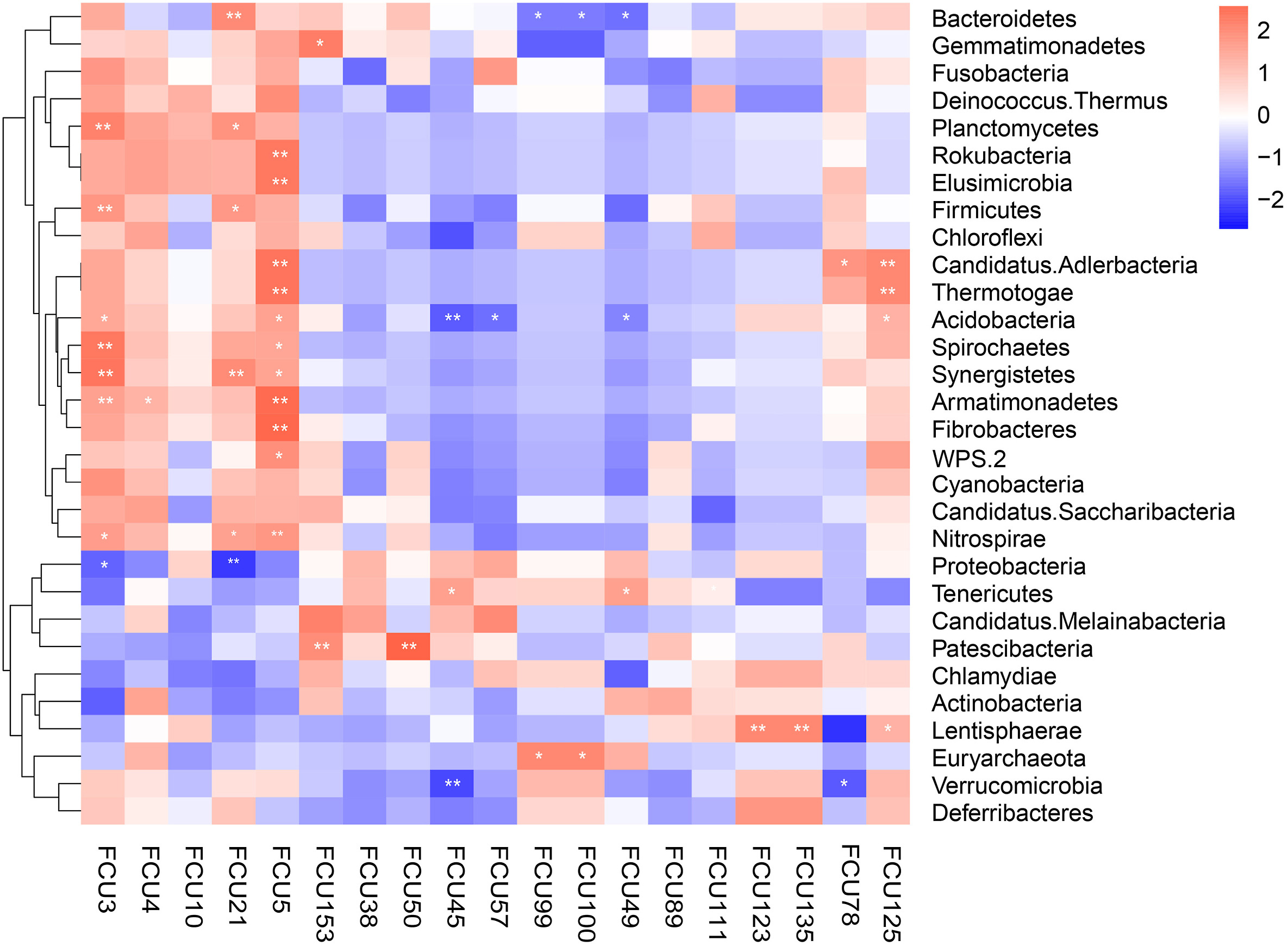
We assessed the correlation between FCUs with the gut microbiota. A Spearman correlation between relative abundances of FCUs and microbial relative abundances yielded two FCU-microbe clusters (Fig. 6). Among these taxa, Firmicutes showed positive correlation with FCU3, FCU21, while Proteobacteria showed negative correlation with FCU3, FCU5 and Bacteroidetes with FCU99, FCU100, FCU49 (p < 0.01 or p < 0.05). In addition, some lower abundance phyla including Planctomycetes, Synergistetes, Armatimonadetes, Nitrospirae were strongly positively correlated with FCU3, FCU4, FCU21, FCU5, whereas they were negatively correlated with FCU45, FCU57, FCU49, FCU123.
Several gut-associated taxa were defined as "core microbiota", the phyla Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes were found in all our samples and are often found in gut habitats across a variety of wild birds (Bodawatta et al., 2018; Grond et al., 2018; Fu et al., 2020). These core microbiota may play an important role in food digestion, nutrient absorption, metabolism, and immunity of wild birds. For example, Firmicutes and Bacteroidetes contribute to the digestion of food sources of proteins, fats, and carbohydrates (Thomas et al., 2011; Cho and Lee, 2020). This seems to be related to the energy accumulation required by the five flycatchers for their altitude migrations in spring (Goulão et al., 2015, Zhang et al., 2015). Proteobacteria, a highly taxonomic and functionally diverse group of bacteria, are related to the functional variability of the gut (Bradley and Pollard, 2017; Grond et al., 2018). Actinobacteria play an important role in regulating intestinal permeability, immune system, and metabolism (Binda et al., 2018). While certain lineages of phyla Proteobacteria, Actinobacteria, Bacteroidales, and Fusobacteria, prevail in the gut of wild birds (Wang et al., 2021a), they are strongly reduced or absent in poultry (Pourabedin and Zhao, 2015). In addition, the diversity of genus-level taxa is also higher in wild birds in our study compared to poultry and much of the community is made up by more uncommon genera (< 1%). A previous study has shown that the random colonization by surrounding environmental bacteria is assumed to be a key reason for a high variation in the intestinal microbiota (Stanley et al., 2013). The environmental conditions of wild bird habitats are complex, including diet, water, soil, nesting environments, and social interactions. The diverse microbes in these environments may become potential sources of inoculation for gut microbiota. These important microbes might be passed down from generation to generation by interaction (Chen et al., 2020), or spread across species due to the closeness of foraging sites (Wang et al., 2021b), together forming the core microbial phyla of these five flycatchers.
Previous studies on birds have provided support for the effect of diet on interspecific variation, including insectivorous, herbivorous, and omnivores etc (Hird et al., 2015; Bodawatta et al., 2018; Capunitan et al., 2020). However, most of these studies use indirect dietary data and dietary data based on literature searches, mostly with limitations. In wild individuals, simple food classification variables often do not accurately reflect species' diets because in nature, most individuals consume a mixture of foods rather than specializing on single items. Thus, to identify and understand the actual diet of flycatchers reported as exclusive insectivorous, we performed a microscopic analysis of their gastric contents. Interestingly, our findings showed that the diet of the five bird species clearly differed in the two traits that we examined: (1) the composition of major food types, (2) the diversity of diet. These finding are complementary to the current knowledge of dietary features of wild flycatchers. Moreover, our research further shows that although the five flycatchers had the same core phyla, there were interspecific differences in their gut microbiota composition, and secondly, we detected significant correlations between multiple FCUs and gut microbiota phylum-level abundance. This seems to indicate that food composition is highly influential on the gut microbiota. Several factors affect the composition and evolution of the avian gut microbiota (Bodawatta et al., 2022a) and diet composition is considered the main factor contributing to the diversity of the gut microbiota in wild vertebrates (Hird et al., 2015; Miyake et al., 2015, Xiao et al., 2021). We found that the relative abundance of Bacteroidetes was positively associated with the proportion of insects in food (Appendix Fig. S7). This coincides with the function of Bacteroidetes involved in the metabolism of high-protein foods in the gut (Thomas et al., 2011). For the Blue-fronted Redstart and the Himalayan Bluetail, which showed a certain preference for fruit, the abundance of Bacteroidetes was therefore significantly lower than that of the other three flycatchers that would select high-protein insects as their only food components. It is worth noting that, FCUs appeared to show significant associations with many less abundant phyla, including Gemmatimonadetes, Chloroflexi, Tenericutes, Verrucomicrobia, Planctomycetota, etc. We consider that it is probably related to the ingestion of microbiota on/in the surface of the food by the host (Laparra & Sanz, 2010; Li et al., 2017) and inoculation with potential environmental microorganisms (e.g. lichens and bryophytes) (Buckley et al., 2006). Gemmatimonadetes, Chloroflexi, etc. are frequently reported in the gut of insects (Huang et al., 2021), and previous study has shown that the gut microbiota of the Prussian carp is very similar to that of the chironomid larvae it eats (Kashinskaya et al., 2015). Planctomycetota is considered an environmental organism and is often observed in association with lichens and bryophytes (Buckley et al., 2006). In addition to the above two explanations for the potential sources of gut microbiota, the interspecies-specific dietary habits that allow bacteria with higher energy collection and storage efficiencies to accumulate in the gut are also important reasons for diet influences the gut microbial composition (Xiang et al., 2021).
Besides, evolutionary history, physiology and genetic factors including host immune genes, pedigree, etc., and behavioral factors, including parental microbial transfer and social interaction, all influence the establishment and composition of the gut microbiota, and also the gut microbes respond to internal and external environmental changes, parasitic infections, diseases, etc., but this is beyond the scope of our present study. Future research could explore these aspects.
The five flycatchers in our study are similar in size, morphology, and feeding type and occupy the same habitats, but direct evidence of competition for resources between them is lacking. In our findings, insects were the main component in their food. However, differing preferences for insect species selection resulted in significant differences in the diet overall. Dietary partitioning is one axis by which sympatric species avoid competition, and is typically documented by cataloging diets to show minimal overlap, describing the morphological traits that facilitate dietary specialization (Greene et al., 2020). Niche partitioning is an important mechanism for allowing ecologically similar species to coexist because resource partitioning facilitates sympatric coexistence (Schoener, 1974; Grevé et al., 2019). Gut microbiota — the consortia of microorganisms, including bacteria, archaea, fungi, and protists that inhabit animal gastrointestinal tracts — represent an underappreciated means of partitioning resources and they are metabolically versatile. Five flycatchers species exhibited similar gene functions at KEGG level 2, but more differences in functional enrichment of gut microbiota at KEGG level 3. This finite difference in metabolic pathways may be a strong fit between gut microbes and nutritional ecology in response to the availability of food resources (Felice et al., 2019). On the other hand, via their role in digesting substrates unavailable to hosts, can help animals expand their dietary options. Therefore, the study of the gut microbial composition of sympatric species will further increase our understanding of ecological niche differentiation.
This study describes the composition, diversity, and function of gut microbiota in five sympatric flycatchers. Five species of birds shared dominant phyla of Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes, and also have their own core and unique microbial communities. Although all are insectivorous birds, they have different preferences for food choices. Diet is an important factor influencing the composition of gut microbes. Microbes on/inside food, and microbes in the ingestion environment are all potential sources of inoculum for host gut microbes. Gut microbiota may represent an underappreciated way of resource allocation under dietary differentiation and contribute to our understanding of niche differentiation. Importantly, the correlations of gut microbiota composition and species, gut microbiota composition and food exhibited moderate effects size. Testing more samples and even more species in later experiments may show significant effects. Future studies should focus on linking our described diet-gut microbial composition associations to host adaptations to determine the positive role of the gut microbiota in host ecological niche differentiation.
Zhehan Dong: Conceptualization, Methodology, Investigation, Data Curation, Formal Analysis, Visualization, Writing - Original Draft, Writing - Review & Editing; Shangmingyu Zhang: Investigation, Data Curation, Writing - Review & Editing; Yuwen Cheng: Investigation, Data Curation, Visualization; Xingcheng He: Conceptualization, Writing - Review & Editing; Ian Haase: Writing - Review & Editing; Yi Liang: Investigation; Yong Jiang: Investigation; Yongjie Wu: Conceptualization, Resources, Funding Acquisition, Project Administration, Supervision, Validation, Writing - Review & Editing. All authors read and approved the final manuscript.
The authors declared that they have no competing interest in this work.
We thank Zhengwei Liu, Xiaofeng Zheng, Kaize Feng, Yanni Zhang, Kexin Peng for their kind assistance with fieldwork and data collection. We thank Faming Liu and Quanxiao Lan at Gongga Alpine Ecosystem Observation and Research Station for their kind assistance with the fieldwork. We thank Nanjiong Pang and Dongrui Li for their advice and assistance during the writing of the manuscript. This study was financially supported by The Second Tibetan Plateau Scientific Expedition and Research Program (No. 2019QZKK0501) and National Natural Science Foundation of China (No. 31772478, 31501851).
All animal research procedures strictly complied with the P.R. China Legislation on the Use and Care of Laboratory Animals and were approved by the Animal Care Review Committee, College of Life Sciences, Sichuan University, China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.avrs.2022.100050.
| 1. | Jane M. Reid, A. Bradley Duthie, Matthew E. Wolak, et al. Demographic mechanisms of inbreeding adjustment through extra-pair reproduction. Journal of Animal Ecology, 2015. DOI:10.1111/1365-2656.12340 |
| 2. | Sylvain Losdat, Peter Arcese, Jane M. Reid, et al. Double decomposition: decomposing the variance in subcomponents of male extra-pair reproductive success. Journal of Animal Ecology, 2015. DOI:10.1111/1365-2656.12389 |
| Common name | Scientific name | Intestinal tract Reps | Intestinal tract Reps ID | Stomach contents Reps | Stomach contents Reps ID |
| Rufous-gorgeted Flycatcher | Ficedula strophiata | 3 | RF223, RF224, RF242 | 6 | RF195, RF223, RF224, RF227, RF242, RF256 |
| Daurian Redstart | Phoenicurus auroreus | 5 | DR63, DR65, DR87, DR91, DR122 | 6 | DR63, DR65, DR87, DR116, DR91, DR122 |
| Rufous-bellied Niltava | Niltava sundara | 4 | RN197, RN207, RN253, RN261 | 2 | RN207, RN253 |
| Blue-fronted Redstart | Phoenicurus frontalis | 3 | BR24, BR32, BR33 | 8 | BR18, BR24, BR27, BR28, BR32, BR36, BR94, BR98 |
| Himalayan Bluetail | Tarsiger rufilatus | 3 | HB45, HB92, HB96 | 5 | HB30, HB45, HB76, HB92, HB96 |
| Intestinal tract Reps were the number of intestinal tract sample replicates, and intestinal tract Reps ID corresponds to the individual number of the bird. Stomach contents Reps were the number of stomach contents sample replicates, and stomach contents Reps ID corresponds to the individual number of the bird. | |||||
| Rufous-gorgeted Flycatcher | Daurian Redstart | Blue-fronted Redstart | Himalayan Bluetail | Rufous-bellied Niltava | |
| Observation | 41 | 69 | 38 | 33 | 18 |
| Shannon index | 2.68 | 3.03 | 2.91 | 0.90 | 2.47 |
| Observation was the number of FCUs with the same morphological characteristics and same traits that we actually counted in each species, Shannon index was used to demonstrate the diversity of the stomach contents, the higher value means the richer stomach contents. | |||||