| Citation: | Wenjuan Wang, Yafang Wang, Qing Chen, Huifang Ding. 2023: Effects of diet shift on the gut microbiota of the critically endangered Siberian Crane. Avian Research, 14(1): 100108. DOI: 10.1016/j.avrs.2023.100108 |
Wetlands worldwide have suffered from serious degradation and transformation, leading to waterbirds increasingly dependent on agricultural fields for feeding. Although gut microbiota is an essential component of host health, the impacts of agricultural feeding on gut microbial community and pathogen transmission remain poorly understood. To fill this knowledge gap, we used 16S rRNA sequencing to characterize the fecal bacterial community of the Siberian Crane (Grus leucogeranus), a Critically Endangered species, that recently has shifted its foraging from largely Vallisneria tubers in Poyang Lake natural wetlands to crops (i.e., rice seeds and lotus rhizomes) in agricultural fields. We compared the bacterial communities between tuber foraging cranes and crop foraging cranes. Our results indicate that diet shift greatly modified the gut microbiota diversity, composition and function. Crop foraging cranes had higher microbiota diversity than tuber foraging cranes. The alteration in microbiota composition and function were correlated with change in food nutrition. Tuber (i.e., high in fiber) foraging cranes were enriched in Clostridiaceae with fiber digestion ability, and crop (i.e., high in carbohydrate) foraging cranes were enriched in bacterial taxa and functions related to carbohydrate metabolism. The flexibility of gut microbiota might enhance Siberian Cranes' ability to adapt to novel diet and environment. However, many enriched families in crop foraging cranes were pathogenic bacteria, which might increase the susceptibility of cranes to pathogenic infection. Special caution should be taken to agricultural feeding waterbirds in Asia, where the widespread poultry-keeping in over-harvested rice fields might increase the transmission probability of pathogenetic bacteria among wild birds, domestic poultry and humans.
Due to intensified human activities and global climate changes, wetlands worldwide have suffered from serious degradation and loss (Davidson, 2014; Phillips et al., 2016), particularly in China (Meng et al., 2017; Zhang et al., 2017). Wetland deterioration has driven many waterbirds to become more dependent on agricultural habitats for feeding (Czech and Parsons, 2002; Fox et al., 2017; Hou et al., 2020). Agricultural feeding has mixed effects. Some bird populations have collapsed because of agricultural intensification, whereas others have benefited from the transition from natural wetlands to agricultural fields (Amano, 2009; Fox and Abraham, 2017; Hemminger et al., 2022). Agricultural feeding can influence various aspects of bird lives, including activity budgets, food intake rate, and fat accumulation (Fox and Abraham, 2017). It also can have fitness consequences by influencing breeding success and abundance (Fox and Abraham, 2017; Fox and Madsen, 2017). Although gut microbiota is an essential component of host health (Hall et al., 2017; Grond et al., 2018), the impacts of agricultural feeding on gut microbiome and possible downstream effects on waterbird fitness remain poorly understood.
Birds harbor a community of microbes, with densities as high as 1011 CFU/g in the hindgut (Barnes, 1972). Gut microbiome plays an important role in host metabolism (Li et al., 2008), behavior (Ezenwa et al., 2012), development (Gilbert et al., 2015) and immune function (Hooper et al., 2012). The characteristics of gut microbiota are primarily shaped by host taxonomy (Ley et al., 2008; Hird et al., 2015). It is also greatly influenced by environmental factors, of which diet seems to be the most important factor (Ley et al., 2008; Muegge et al., 2011; Waite and Taylor, 2014). Diet shifts have been reported to lead to modification of gut microbial compositions in humans (Clarke et al., 2014; David et al., 2014), other mammals (Muegge et al., 2011; Li et al., 2020b), and birds (Davidson et al., 2020; Teyssier et al., 2020). Different food items contain different nutrients, some of which can modulate gut microbiota composition, such as protein (Clarke et al., 2014), fiber (David et al., 2014; Tang et al., 2021), fat (Fava et al., 2013) and secondary compounds (Li et al., 2019a). Compared to natural plants, agricultural crops generally contain higher protein, fat and energy, but lower fiber and ash (Fox and Abraham, 2017). These differences may alter the gut microbiome after hosts shift from eating wild plants to crops.
Changes in gut microbiota composition usually are accompanied by changes in of microbiota function (Li et al., 2020a; Teyssier et al., 2020). The change degree of microbiota function appeared to be more conserved than microbiota composition as different bacteria may have similar functions (Li et al., 2021). The modulations of gut microbiota composition and function can have downstream impacts on host health and fitness (West et al., 2019; Li et al., 2020a, 2020b). Given the important role of gut microbiota in host health, it is important to consider gut microbiota research in threatened species conservation (West et al., 2019).
Poyang Lake, the largest freshwater lake in China, is a globally important wintering ground for waterbirds in the East Asian–Australasian Flyway (Cao et al., 2010; Wang et al., 2017). It is the most important wintering ground for the IUCN Critically Endangered Siberian Crane (Grus leucogeranus), harboring 98% of its global population (BirdLife International, 2022). The Siberian Crane is the most aquatic of cranes, relying on wetlands for nesting, feeding and roosting (Harris and Mirande, 2013; Hemminger et al., 2022). It traditionally feeds in shallow waters on tubers of Vallisneria spp., the dominant submerged macrophyte at Poyang Lake (Wu et al., 2013; Hou et al., 2020).
However, the density and biomass of submerged macrophyte at Poyang Lake have declined seriously in the recent decades due to floods and droughts, extensive aquaculture, declining water quality, and sand mining (Hu and Lin, 2019; Hou et al., 2020; Li et al., 2020a). The submerged macrophyte degradation has resulted in changes in diet and foraging habitat of waterbirds that formerly specialized on Vallisneria tubers, including Siberian Cranes (Jia et al., 2013; Hou et al., 2020), White-naped Cranes (Grus vipio; Hou et al., 2021), and Tundra Swans (Cygnus columbianus bewickii; Li et al., 2013). Siberian Cranes have shifted from eating Vallisneria tubers in shallow waters to eating rice seeds and lotus rhizomes in rice paddies and lotus ponds (Hou et al., 2019, 2021). These changes were accompanied by a decrease in Siberian Crane foraging time (Shao et al., 2018) and a reduction of dietary niche overlap among crane species (Hou et al., 2021). Whether diet shift affected gut microbiota and then the health condition of Siberian Cranes remain unknown.
Here, we assessed the effects of Siberian Cranes shifting from natural wetlands to agricultural feeding on their gut microbiota composition, diversity and function, and on the transmission of pathogenic bacteria. We first compared the composition, diversity and function of gut microbial communities of crop foraging cranes (i.e., cranes primarily fed on rice seeds and lotus rhizomes in agricultural fields) and tuber foraging cranes (i.e., cranes primarily fed on Vallisneria tubers in natural wetlands). We hypothesized that different nutrient compositions between Vallisneria tubers and agricultural crops would alter gut microbiota composition, diversity and function of Siberian Cranes. After that, we compared the abundance of pathogenic bacteria between crop foraging cranes and tuber foraging cranes. Numerous free-ranging domestic fowl have been raised by traditional husbandry in rice paddies around Poyang Lake (Choi et al., 2016; Xing and Wang, 2016). Agricultural feeding would increase the chances of wild birds contacting with domestic fowl and humans, which, in turn, would lead to higher risk of transmission of pathogens among wild birds, domestic fowl and humans (Muzaffar et al., 2010; Prosser et al., 2016; Fu et al., 2020). Therefore, we expected a higher abundance of pathogenic bacteria in gut microbiota of crop foraging cranes than tuber foraging cranes. Our study could improve our understanding of the impacts of the increasingly occurred agricultural feeding on gut microbiome, health and fitness of birds, and the adaptation mechanism of birds to diet and environment changes.
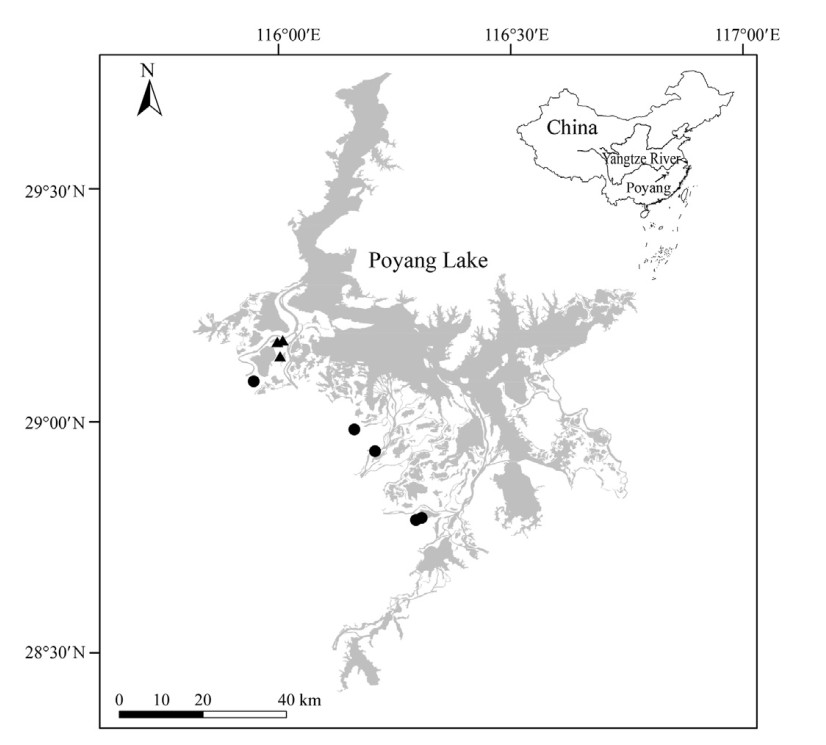
Poyang Lake (Fig. 1), located on the south bank of the Yangtze River, is a Ramsar site and a globally important wintering ground for waterbirds in the East Asian-Australasian Flyway (Cao et al., 2010; Wang et al., 2017). It harbors an average of 426, 000 wintering waterbirds of 111 species (Li et al., 2019b). Poyang Lake is one of only two lakes remaining that are freely connected with the Yangtze River. There are sluices built at the outlet of other lakes in the Yangtze River floodplain. Driven by local precipitation and the upstream Yangtze River, the water level and inundation area of Poyang Lake vary seasonally (Feng et al., 2012). During the wet season (i.e., April–September), the water level increases to about 16.73 m (Dai et al., 2015), and the inundation area expands to > 3000 km2 (Feng et al., 2012). During the dry season (i.e., October–March), the water level declines to about 9.85 m (Dai et al., 2015), and the inundation area shrinks to < 1000 km2 (Feng et al., 2012). The dynamic hydrology is an important factor leading to the high biodiversity of Poyang Lake (Barzen, 2012). Flood pluses during the wet season bring nutrients and organic matter to Poyang Lake, promoting the growth of aquatic organisms, and water recession during the dry season exposes rich feeding areas for numerous waterbirds (Barzen, 2012).
Due to the decline of Vallisneria tubers, we did not collect enough tubers for nutritional composition determination; thus, we used the nutritional data in Jia (2013). We collected rice seeds from paddy fields and lotus rhizomes from lotus ponds used by Siberian Cranes. Samples were oven-dried at 60 ℃ to constant weight, and then ground to powder. Two samples of rice seeds and three samples of lotus rhizomes with dry weights > 100 g were used for nutritional evaluation. The contents of starch, fiber, protein, fat, and ash were determined according to the Chinese National Standards GB 5009.9-2016, GB/T 5009.10-2003, GB 5009.5-2010, GB 5009.6-2016, and LY/T 1268-1999, respectively (China National Forestry Industry Standards, 1999; China National Food Safety Standard, 2003, 2010, 2016a, 2016b). Sugar was determined by anthrone colorimetry. The nutritional determination methods were similar between Jia (2013) and our study. Nutritional measurements were conducted by the Beijing Sino-Instrument and Test Technology Company Limited (Beijing, China). Carbohydrate content was calculated by adding the proportions of starch, sugar and fiber.
Our previous study indicated that winter 2017 was a year with low Vallisneria tuber abundance, while winter 2018 was a year with high Vallisneria tuber abundance (Hou et al., 2021). Siberian Cranes primarily fed on rice seeds and lotus rhizomes in rice paddies and lotus ponds in winter 2017 because of food shortage in natural wetlands (Hou et al., 2019, 2021). When the abundance of Vallisneria tuber rebounded in winter 2018, Siberian Cranes moved back to natural wetlands to feed on Vallisneria tubers and Polygonum criopolitanum rhizomes (Hou et al., 2021). Similarly, Zhong (2020) recorded 1222 Siberian Cranes in the seven transects of rice paddies around Poyang Lake in winter 2017, but no Siberian Cranes in winter 2018. In this study, we collected Siberian Crane feces from rice paddies and lotus ponds in winter 2017, and from natural wetlands in winter 2018. Our field observations indicated that Siberian Cranes moved frequently between rice paddies and lotus ponds during low tuber periods; thus, fecal samples from the two habitats were combined to represent agricultural feeding.
As the Siberian Crane is a Critically Endangered species, we used noninvasive method to collect fecal samples (Darimont et al., 2008). To ensure the species origin of fecal samples, we collected samples from monospecific groups of Siberian Cranes. When we saw Siberian Cranes, we waited until they moved to reduce interference. Soon after they left, we collected fresh feces with sterilized tweezers. To avoid the contamination from soils, we only collected partial feces that did not contact soil. Samples were stored in sterile 10 mL collection tubes in liquid nitrogen in the field until transferred to a −80 ℃ refrigerator in the laboratory. To reduce the probability of collecting multiple samples from the same individuals, fecal samples were collected at a distance of at least 5 m from one anther (Hou et al. 2021).
DNA was extracted from about 1g fecal sample using QIAamp PowerFecal Pro DNA Kit (Qiagen, Germany). We amplified the variable V3–V4 region of bacterial 16S rRNA using primers 341F (5′-CCT ACG GGN GGC WGC AG-3′) and 805R (5′-GAC TAC HVG GGT ATC TAA TCC-3′) with Illumina adapter sequences on the 5′ end (Herlemann et al., 2011). The PCR amplification procedure was as follows: denaturation at 94 ℃ for 2 min, followed by 27 cycles at 94 ℃ for 30 s, 55 ℃ for 30 s, and 72 ℃ for 1 min, and a final 10 min at 72 ℃. Three PCR replicates were performed for each sample to reduce amplification bias. DNA concentration and purity was checked on a 1% agarose gel. Samples with a clear band were mixed and purified using the Agencourt AMpure XP (Beckman Coulter, USA). Libraries were prepared and sequenced on the Illumina MiSeq 2 × 250 platform (Illumina, USA) by the Shanghai Genesky Biotechnologies Inc. (Shanghai, China). Sequence data were submitted to the NCBI Sequence Read Archive under the accession numbers PRJNA974217.
Adapter sequences and primers of raw reads were trimmed using the cutadapt plugin of QIIME2 (Bolyen et al., 2019). The DADA2 algorithm of QIIME2 (Callahan et al., 2016) was used for quality control and to identify amplicon sequence variants (ASVs). Taxonomic assignments of ASV were performed using RDP Classifier (Wang et al., 2007) with a confidence threshold of 0.8 against the SILVA 16S rRNA database (Quast et al., 2013). Sequences classified as chloroplast and mitochondria were removed from further analysis. The ASV table was rarefied to the minimum number of reads per sample using R (version 3.5.1; R Core Team, Vienna, Austria) package phyloseq (McMurdie and Holmes, 2013) to avoid the influence of difference in read numbers for subsequent analyses. Sample-based rarefaction curves were built to examine the sufficiency of sequencing depth. The curves were computed by randomly resampling sequences and plotting them against the numbers of ASVs using QIIME2. We also calculated Good's Coverage using R package vegan (Oksanen et al., 2013) to evaluate the sequencing effectiveness.
To compare the microbiota composition between the two foraging groups, we first examined variation in alpha diversity indices, including ASV number, Chao1 estimator, ACE estimator, and Shannon–Wiener diversity. A Wilcoxon rank sum test was used to test the difference in alpha diversity between the two foraging groups. We then examined beta diversity among samples. We conducted Principal Co-ordinates Analysis (PCoA) based on Bray–Curtis and Jaccard distance measures. The Jaccard index compares community similarity based on the presence/absence of ASVs, and the Bray–Curtis index compared community similarity based on ASV abundance. Analysis of similarities (ANOSIM) based on Bray-Curtis distance was used to evaluate if microbial communities were significantly different between the two foraging groups. Metastats analysis was conducted to determine which taxon differed significantly between the two foraging groups based on ASVs with a relative abundance > 0.1%. The R package vegan and phyloseq were used to do the above analyses, and ggplot2 (Wickham, 2009) was used to create graphics.
To explore the differences in gut microbiota function between the two foraging groups, we predicted the 16S rRNA database through PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) analysis. We analyzed the functional information of these sequences through KEGG (Kyoto Encyclopedia of Genes and Genomes) database. We used ANOSIM analysis to assess if the microbial function significantly differed between the two foraging groups. Welch's T-test was used to determine which gene function differed significantly between the two groups. The ANOSIM and T-test analyses were carried out using R.
The starch and protein contents were lower in Vallisneria tuber than in rice seed and lotus rhizome, which also led to a lower proportion of carbohydrate in Vallisneria tuber than in rice seed and lotus rhizome (Table 1). The proportions of fiber and fat in Vallisneria tuber were higher than that in rice seed and lotus rhizome.
| Food item | Starch | Protein | Fiber | Ash | Fat | Carbohydrate |
| Vallisneria tuber | 19.53 (4.92) | 4.24 (0.74) | 1.61 (0.59) | 1.87 (1.19) | 0.29 (0.06) | 20.69 (5.07) |
| Rice seed | 58.60 (4.00) | 6.52 (0.33) | 0.17 (0.06) | 3.29 (0.21) | 0.08 (0.01) | 59.28 (4.34) |
| Lotus rhizome | 56.43 (6.26) | 8.00 (0.49) | 0.37 (0.24) | 4.57 (0.23) | 0.07 (0.01) | 57.49 (6.98) |
Overall, we collected a total of 44 fecal samples, including 19 samples from natural wetlands and 25 samples from agriculture fields (i.e., 13 samples from rice paddies and 12 samples from lotus ponds; Fig. 1 and Table S1). For each foraging habitat (i.e., natural wetlands, rice paddies and lotus ponds), we collected feces from 2 to 4 sites to reflect spatial variation of gut microbial communities. Illumina sequencing of the 44 fecal samples yielded a total of 6, 954, 262 raw reads (mean = 160, 880 ± 66, 298 reads per sample). After quality control, each sample contained an average of 126, 786 ± 59, 978 high-quality reads. Sample-based rarefaction curves plateaued in most cases as the number of sequences increased, indicating that our sequencing depth was adequate to capture the gut microbiota diversity (Appendix Fig. S1). The average value of Good's Coverage was 0.999 ± 0.001, also indicating that the majority of bacterial diversity was captured. A total of 9104 ASVs were identified (mean = 356 ± 280 ASVs per sample). The ASV table was rarefied to 52, 618 reads per sample for statistical analyses, which reduced the total number of ASVs to 8109 (328 ± 247 ASVs per sample). In total, 46 phyla, 112 classes, 172 orders, 213 families, and 351 genera were identified.
Overall, 99.69% filtered sequence reads were assigned to order level. About 89.21% reads were assigned to family level in crop foraging cranes, but only 51.08% reads were assigned to family level in tuber foraging cranes. The unassigned reads primarily came from order Clostridiales. The most dominant phyla across all samples were Firmicutes (85.71% ± 16.01), Proteobacteria (10.31% ± 13.48) and Actinobacteria (2.32% ± 3.84; Fig. 2). The most dominant families across all samples were Lactobacillaceae (42.99% ± 28.06) and Clostridiaceae (9.53% ± 13.63). Lactobacillaceae was significantly higher in crop foraging cranes (56.40%) than in tuber foraging cranes (26.11%; P = 0.001), while Clostridiaceae was significantly higher in tuber foraging cranes (17.54%) than in crop foraging cranes (3.08%; P = 0.002; Table 2). Tuber foraging cranes had significantly higher relative abundance only in family Clostridiaceae, while crop foraging cranes had significantly higher abundances in 24 families (Table 2), half of them have been reported to be pathogenic bacteria (see Discussion for details).
| Taxon | Tuber_mean (%) | Tuber_SD (%) | Crop_mean (%) | Crop_SD (%) | P |
| Lactobacillaceae | 26.11 | 4.69 | 56.40 | 5.27 | 0.001 |
| Clostridiaceae | 17.54 | 3.83 | 3.08 | 1.15 | 0.002 |
| Enterobacteriaceae | 0.02 | 0.01 | 8.71 | 2.77 | 0.001 |
| Streptococcaceae | 0.08 | 0.04 | 2.31 | 0.86 | 0.001 |
| Rhizobiaceae | 0.02 | 0.01 | 1.39 | 0.37 | 0.001 |
| Micrococcaceae | 0.07 | 0.06 | 0.90 | 0.27 | 0.002 |
| Microbacteriaceae | 0.00 | 0.00 | 0.68 | 0.20 | 0.001 |
| Hyphomicrobiaceae | 0.19 | 0.07 | 0.54 | 0.15 | 0.028 |
| Fusobacteriaceae | 0.07 | 0.03 | 0.45 | 0.21 | 0.044 |
| Streptomycetaceae | 0.03 | 0.01 | 0.44 | 0.19 | 0.007 |
| Methylobacteriaceae | 0.01 | 0.00 | 0.42 | 0.13 | 0.001 |
| Corynebacteriaceae | 0.04 | 0.04 | 0.37 | 0.12 | 0.017 |
| Paenibacillaceae | 0.05 | 0.02 | 0.32 | 0.11 | 0.007 |
| Mycobacteriaceae | 0.01 | 0.01 | 0.26 | 0.06 | 0.001 |
| Phyllobacteriaceae | 0.00 | 0.00 | 0.26 | 0.10 | 0.003 |
| Staphylococcaceae | 0.00 | 0.00 | 0.24 | 0.12 | 0.001 |
| Succinivibrionaceae | 0.01 | 0.01 | 0.22 | 0.11 | 0.029 |
| Leuconostocaceae | 0.00 | 0.00 | 0.17 | 0.10 | 0.049 |
| Nocardioidaceae | 0.02 | 0.02 | 0.16 | 0.06 | 0.044 |
| Thermogemmatisporaceae | 0.00 | 0.00 | 0.16 | 0.08 | 0.002 |
| Isosphaeraceae | 0.00 | 0.00 | 0.13 | 0.06 | 0.001 |
| Alcaligenaceae | 0.01 | 0.00 | 0.12 | 0.04 | 0.007 |
| Nocardiaceae | 0.01 | 0.01 | 0.11 | 0.04 | 0.006 |
| Brucellaceae | 0.00 | 0.00 | 0.11 | 0.03 | 0.001 |
| Aurantimonadaceae | 0.00 | 0.00 | 0.10 | 0.04 | 0.001 |
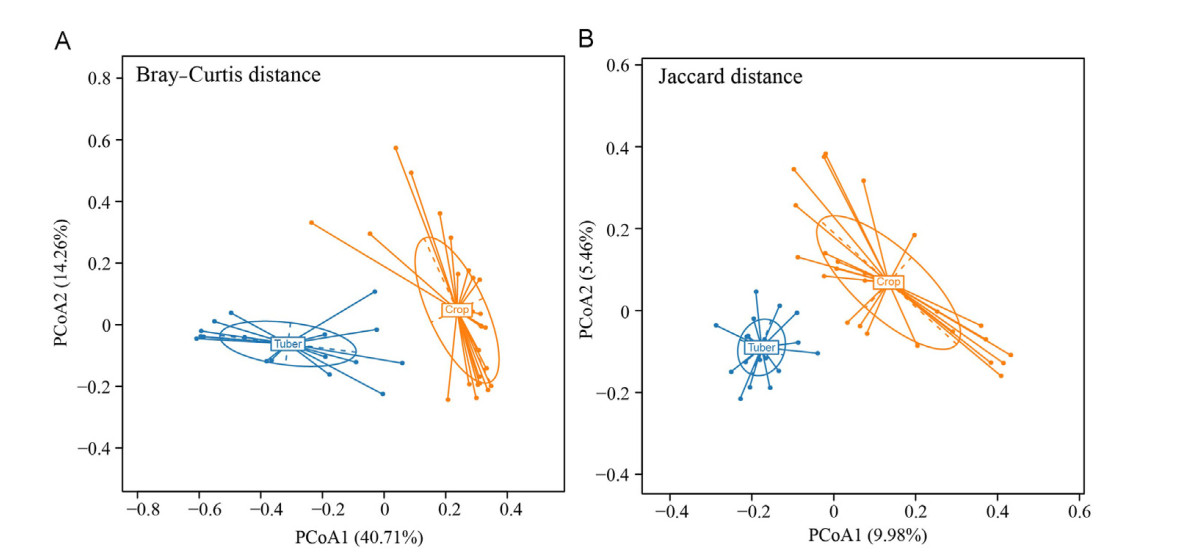
The ASV number (W = 329.5, P = 0.030), Chao1 estimator (W = 146, P = 0.030), ACE (W = 326, P = 0.036), and Shannon–Wiener diversity (W = 325, P = 0.038) were significantly lower in tuber foraging cranes than that in crop foraging cranes (Fig. 3). The ANOSIM analysis indicated that the gut microbiota composition differed significantly between the two foraging groups (R = 0.63, P = 0.000). PCoA based on Bray–Curtis distance indicated that tuber foraging cranes clustered apart from crop foraging cranes (Fig. 4A). PCoA1 and PCoA2 explained 40.71% and 14.26% of the total variation, respectively. PCoA based on Jaccard distance also separated the two foraging groups (Fig. 4B).
The ANOSIM analysis indicated that the KEGG pathway profile of tuber foraging cranes diverged significantly from that of crop foraging cranes (R = 0.21, P = 0.001). The diet shift significantly altered the abundance of 112 pathways. Among the top 30 high abundant pathways, 23 pathways involved metabolism, such as carbohydrate metabolism, amino acid metabolism, metabolism of terpenoids and polyketides, and metabolism of cofactors and vitamins (Fig. 5). There were 15 pathways upregulated and 15 pathways downregulated in crop foraging cranes. Seven pathways upregulated in crop foraging cranes were related to carbohydrate metabolism, including fatty acid biosynthesis, glycolysis/gluconeogenesis, pyruvate metabolism, C5-Branched dibasic acid metabolism, citrate cycle, fructose and mannose metabolism, and amino sugar and nucleotide sugar metabolism.
To the best of our knowledge, we evaluated, for the first time, the impacts of agricultural feeding on the gut microbiome of waterbirds. Our study indicates that the Siberian Crane, a Critically Endangered species and a flagship species of wetland conservation, shifting its foraging focus from Vallisneria tubers to rice seeds and lotus rhizomes, was accompanied by changes in gut microbiota diversity, composition and function. Agricultural feeding was associated with higher microbiota diversity, changes in taxonomic composition, and changes in functional composition. Agricultural feeding was also associated with an increase in the abundance of pathogenic bacteria. Given the important role of gut microbiome in host health, it is important to consider gut microbiome when we evaluate the impacts of the increasingly occurred agricultural feeding on waterbirds.
The gut microbiota composition and function of crop foraging cranes differed significantly from tuber foraging cranes. Diet-induced alterations in gut microbiota composition and function have been documented in other animals. Extreme short-term diets had a pronounced effect on human microbiota (David et al., 2014). Seasonal variations in diets have been reported to result in seasonal reconfiguration of gut microbiota in birds (Michel et al., 2018; Góngora et al., 2021) and other mammals (Amato et al., 2015; Wu et al., 2017; Hicks et al., 2018). Migratory birds exposed to variable food resources and environments during seasonal migration also exhibited seasonal shifts in gut microbiota (Lewis et al., 2017; Zhang et al., 2020). Our study indicated that birds shifting from eating wild plants to crops can also have pronounced impacts on gut microbiota.
The gut microbiota composition and function of Siberian Cranes showed an adaptation or adjustment to their food nutrition. Vallisneria tuber contains higher abundance of fiber than rice seed and lotus rhizome. Correspondingly, Clostridiaceae, which contains taxa known to digest cellulose (Van Dyke and McCarthy, 2002; Burrell et al., 2004), was higher in tuber foraging cranes. Higher dietary fiber intake has also been reported to stimulate the increase of Clostridiaceae in the guts of humans (Medawar et al., 2021) and mice (Nagy-Szakal et al., 2013). Rice seed and lotus rhizome contain a higher proportion of carbohydrate than Vallisneria tubers. Accordingly, Lactobacillaceae, which has been reported to play a major role in fermenting carbohydrate to produce lactic acid (Pfeiler and Klaenhammer, 2007), was enriched in crop foraging cranes. Moreover, seven out of the 15 pathways upregulated in crop foraging cranes were related to carbohydrate metabolism. Similarly, Common Cranes (Grus grus), Great Bustards (Otis tarda dybowskii) and Common Coots (Fulica atra) feeding on wheat high in carbohydrate also showed enrichment in the carbohydrate metabolic pathways (Lu et al., 2022). The modifications of bacterial composition and pathways corresponding to food nutrition suggest that microbiota change might enhance the ability of Siberian Cranes to digest foods with different nutrient compositions.
Like Siberian Cranes, alterations in gut microbiome in response to diet composition and nutritional demands have been reported in several vertebrates (David et al., 2014; Amato et al., 2015; Teyssier et al., 2020; Baniel et al., 2021). For example, cellulolytic/fermentative bacteria increased in the gut of Ethiopian Geladas (Theropithecus gelada) during wetter periods, when geladas primarily fed on cellulose rich grass, while amylolytic and methanogenic bacteria increased during drier periods, when geladas incorporated more starch and lignified food into their diet (Baniel et al., 2021). Urban House Sparrows (Passer domesticus) fed with a rural diet (i.e., high fiber diet) showed an increase in the abundance of two families of Firmicutes (Enterococcaceae and Staphylococaceae), a phylum associated with metabolism of dietary plant polysaccharides (Teyssier et al., 2020). Our study, in combination with these previous researches, suggests that gut microbiota represents an important source of metabolic flexibility for hosts, and might play an essential role in facilitating host acclimatization to diet and environmental changes (Alberdi et al., 2016; Macke et al., 2017; Baniel et al., 2021).
Crop foraging cranes had higher alpha diversity than tuber foraging cranes. High gut microbiota diversity generally is considered beneficial to hosts (Le Chatelier et al., 2013). More diverse microbial communities can increase vertebrates' capacity to assimilate nutrients (Le Chatelier et al., 2013), resist pathogen invasion (Ganz et al., 2017), and adapt to environmental changes (Alberdi et al., 2016). Higher gut microbiota diversities have been documented in healthy people compared to unhealthy and old people (Manichanh et al., 2006; Claesson et al., 2012; Le Chatelier et al., 2013), and in avian influenza–negative Mallards (Anas platyrhyncos) compared to avian influenza–positive Mallards (Ganz et al., 2017). Birds occupying high quality habitats also had higher gut microbiota diversity than birds occupying degraded habitats (Fuirst et al., 2018; Knutie et al., 2019; Teyssier et al., 2020). The higher alpha diversity of crop foraging cranes suggests that the microbiota functions might not be negatively affected by agricultural feeding.
Tuber foraging cranes were enriched only in family Clostridiaceae, which went down to genus Clostridium. This genus contains several pathogenic species. C. perfringens is able to cause a wide range of myonecrotic and gastrointestinal diseases in humans and birds (Van Immerseel et al., 2009). Necrotic enteritis, one of the economically important diseases in poultry industry, is caused by C. perfringens infection (Van Immerseel et al., 2009). C. difficile, a leading cause of antibiotic-associated diarrhea and colitis in humans, can also infect poultry and captive ostriches (Struthio camelus; Moono et al., 2016).
Compared to tuber foraging cranes, crop foraging cranes showed increases in much more families, among which twelve families have been reported to be pathogenic bacteria in humans, including Enterobacteriaceae (Chen et al., 2011; Gevers et al., 2014; Salamon et al., 2018), Streptococcaceae (Chen et al., 2011; Le Bastard et al., 2018), Microbacteriaceae (Kobayashi et al., 2015), Mycobacteriaceae (Scanu et al., 2007), Brucellaceae (Franc et al., 2018), Micrococcaceae (Meng et al., 2018; Margiotta et al., 2021), Nocardiaceae (Abreu et al., 2015; Gao et al., 2020), Paenibacillaceae (Fontana et al., 2020), Alcaligenaceae (Bajaj et al., 2012; Pan et al., 2020), Hyphomicrobiaceae (Zhang et al., 2019), Fusobacteriaceae (Bajaj et al., 2012; Gevers et al., 2014), and Nocardioidaceae (Kim et al., 2021; Wang et al., 2022). Among the twelve potential pathogens in humans, five families have also been reported to be potential pathogens in birds. Specifically, Enterobacteriaceae has been reported to be pathogens to Yellow-headed Blackbirds (Xanthocephalus xanthocephalus; Gibbs et al. 2007) and chicken (Wooley et al. 2000). Streptococcaceae was reported to negatively affect the growth of Crested Ibis (Nipponia nippon; Zhu et al., 2021) and ostrich (Videvall et al., 2019). Mycobacteriaceae (e.g., Mycobacterium) can cause avian tuberculosis, one of the most important diseases affecting birds (Dhama et al., 2011). Micrococcaceae has been observed to be enriched in avian influenza–positive mallards compared to avian influenza–negative mallards (Ganz et al., 2017). Nocardiaceae (e.g., Nocardia) can cause lung lesions in Pesquet's Parrots (Psittricbas fulgidus; Long et al., 1983) and Black Crakes (Limnocorax flavirostra; Bacciarini et al. 1999).
The enrichment of pathogenic bacteria in crop foraging cranes might be due to the intermixture between Siberian Cranes, domestic poultry and humans. Keeping poultry in over-harvested rice fields is a widespread poultry production method in Asia (Muzaffar et al., 2010). There are millions of domestic poultry raised by traditional husbandry in rice paddies surrounding Poyang Lake, where they glean waste grain residues in stubble fields (Choi et al., 2016). In the past decades, the degradation of submerged plants has driven tens of thousands of waterbirds (e.g., cranes, geese, and swans) from feeding in natural wetlands to feeding in agricultural fields (Zhong, 2020; Hou et al., 2021). The foraging habitat shift increases the chance of intermixing between wild birds, domestic poultry and humans (Muzaffar et al., 2010; Xiang et al., 2019), leading to an increase in transmission probability of intestinal microbiota among hosts (Grond et al., 2014; Alm et al., 2018; Fu et al., 2020). Previous studies have reported the transmissions of intestinal pathogens between sympatric wintering Hooded Cranes (Grus monacha) and domestic poultry at Shengjin Lake, another lake in the Yangtze River floodplain (Fu et al., 2020; Mahtab et al., 2021). Urbanization-induced increases in human–animal interactions also have been linked to the prevalence of pathogenic bacteria in the guts of Herring Gulls (Larus argentatus; Fuirst et al., 2018) and American White Ibis (Eudocimus albus; Murray et al., 2020). Moreover, the enrichment of pathogenic bacteria in crop foraging cranes might also be because of dense flocks of waterbirds in some agricultural fields. The government of Jiangxi Province, where Poyang Lake is located, has established two food-supply locations for Siberian Cranes to alleviate the negative impacts of food shortage in natural wetlands. One site is rice paddies located at Kangshan County, Shangrao, and another site is lotus ponds and rice paddies located at the Wuxing Farmland, Nanchang. Thousands of Siberian Cranes gathered at the two locations for feeding (Wang et al., 2019; Di et al., 2022). Besides Siberian Cranes, there were numerous Common Cranes, Tundra Swans, Swan Geese (Anser cygnoides), and ducks (Shao et al., 2018; W. Wang, personal observation). The dense flocks might increase the possibility of cross-species transmission of pathogens among birds (van Veelen et al., 2017; Yang and Zhou, 2021), leading to the enrichment of gut pathogens in Siberian Cranes.
The enrichment of pathogenic bacteria in crop foraging cranes suggests that agricultural feeding might increase Siberian Cranes' susceptibility to pathogen infection, potentially threatening their survival. Since these enriched bacteria primarily were recognized as pathogens in humans as well as limited bird species, further in-depth studies should be conducted to determine if the occurrence of these potential pathogenic bacteria would induce disease in Siberian Cranes. To reduce the prevalence of pathogenic bacteria in Siberian Cranes, we suggest two measures. First, keeping free-range poultry in over-harvested rice paddies should be restricted at the key areas of Siberian Cranes. According to previous researches (He et al., 2019; Zhong, 2020) and our field observation, there were several key farmlands where Siberian Cranes were commonly observed, including Kangshan County, Wuxing Farmland, Zhugang Farmland, Henghu Farmland, and Yongxiu County. Restricting free-range poultry farming at these key distribution areas could reduce the chance of transmission of intestinal pathogens among Siberian Cranes, domestic poultry and humans. Second, dense flocks of Siberian Cranes at the two food-supply locations (i.e., Kangshan County and Wuxing Farmland) should be dispersed to more locations. This could be achieved by reducing food abundance at these two areas and adding more food-supply locations.
Our study indicates that Siberian Cranes switched from feeding in natural wetlands to agriculture fields was accompanied by changes in gut microbiota diversity, composition and function. The gut microbiota composition and function showed an adaptation or adjustment to the dietary nutrition of cranes. Crop foraging cranes had a higher gut microbiota diversity than tuber foraging cranes, which might be beneficial to cranes. However, the enrichment of potentially pathogenic bacteria in crop foraging cranes emphasizes the susceptibility of Siberian Cranes to pathogen infection. Special attention should be paid to Asia, where keeping poultry in over-harvested rice fields greatly increases the transmission probability of intestinal pathogenetic bacteria among wild birds, domestic poultry and humans. Moreover, dense flocks of waterbirds in agricultural fields should be avoided to reduce the chance of cross-species transmission of pathogens among wild birds. Due to wetland degradation and transformation worldwide, waterbirds are expected to be increasingly dependent on agricultural fields for feeding (Czech and Parsons, 2002; Fox et al., 2017). Our study illustrates the importance of considering gut microbiome to fully understand and forecast the potential impacts of agricultural feeding on organisms. It also adds to an increasing body of literature suggesting that the gut microbiota might be a key factor influencing host acclimation and adaptation to new environments (Alberdi et al., 2016; Macke et al., 2017; Baniel et al., 2021), which can be particularly important in the current context of global climate changes.
We collected bird fecal samples using a non-invasive method. Our fieldwork was permitted by the Poyang Lake National Nature Reserve and the Nanji Wetland National Nature Reserve.
WW conceived the study, analyzed the data, and wrote the draft. YW, QC, and HD collected fecal samples, conducted the experiments, and analyzed the data. All authors contributed critically to the drafts. All authors read and approved the final manuscript.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
We are grateful to Lanhua Wang, Haiyan Zhou, the Poyang Lake National Nature Reserve, and the Nanji Wetland National Nature Reserve for assisting in sample collection. We thank James D. Fraser from Virginia Tech University for editing the English language of our draft. We also thank the two anonymous reviewers for their helpful comments and suggestions.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.avrs.2023.100108.
| 1. | Geerat J Vermeij, Stewart Edie, Tracey Chapman. When does natural selection take place?. Evolution, 2023. DOI:10.1093/evolut/qpad108 |
| 2. | Ryan B Stephens, Christopher B Burke, Neal Woodman, et al. Skeletal injuries in small mammals: a multispecies assessment of prevalence and location. Journal of Mammalogy, 2018, 99(2): 486. DOI:10.1093/jmammal/gyy020 |
| Food item | Starch | Protein | Fiber | Ash | Fat | Carbohydrate |
| Vallisneria tuber | 19.53 (4.92) | 4.24 (0.74) | 1.61 (0.59) | 1.87 (1.19) | 0.29 (0.06) | 20.69 (5.07) |
| Rice seed | 58.60 (4.00) | 6.52 (0.33) | 0.17 (0.06) | 3.29 (0.21) | 0.08 (0.01) | 59.28 (4.34) |
| Lotus rhizome | 56.43 (6.26) | 8.00 (0.49) | 0.37 (0.24) | 4.57 (0.23) | 0.07 (0.01) | 57.49 (6.98) |
| Taxon | Tuber_mean (%) | Tuber_SD (%) | Crop_mean (%) | Crop_SD (%) | P |
| Lactobacillaceae | 26.11 | 4.69 | 56.40 | 5.27 | 0.001 |
| Clostridiaceae | 17.54 | 3.83 | 3.08 | 1.15 | 0.002 |
| Enterobacteriaceae | 0.02 | 0.01 | 8.71 | 2.77 | 0.001 |
| Streptococcaceae | 0.08 | 0.04 | 2.31 | 0.86 | 0.001 |
| Rhizobiaceae | 0.02 | 0.01 | 1.39 | 0.37 | 0.001 |
| Micrococcaceae | 0.07 | 0.06 | 0.90 | 0.27 | 0.002 |
| Microbacteriaceae | 0.00 | 0.00 | 0.68 | 0.20 | 0.001 |
| Hyphomicrobiaceae | 0.19 | 0.07 | 0.54 | 0.15 | 0.028 |
| Fusobacteriaceae | 0.07 | 0.03 | 0.45 | 0.21 | 0.044 |
| Streptomycetaceae | 0.03 | 0.01 | 0.44 | 0.19 | 0.007 |
| Methylobacteriaceae | 0.01 | 0.00 | 0.42 | 0.13 | 0.001 |
| Corynebacteriaceae | 0.04 | 0.04 | 0.37 | 0.12 | 0.017 |
| Paenibacillaceae | 0.05 | 0.02 | 0.32 | 0.11 | 0.007 |
| Mycobacteriaceae | 0.01 | 0.01 | 0.26 | 0.06 | 0.001 |
| Phyllobacteriaceae | 0.00 | 0.00 | 0.26 | 0.10 | 0.003 |
| Staphylococcaceae | 0.00 | 0.00 | 0.24 | 0.12 | 0.001 |
| Succinivibrionaceae | 0.01 | 0.01 | 0.22 | 0.11 | 0.029 |
| Leuconostocaceae | 0.00 | 0.00 | 0.17 | 0.10 | 0.049 |
| Nocardioidaceae | 0.02 | 0.02 | 0.16 | 0.06 | 0.044 |
| Thermogemmatisporaceae | 0.00 | 0.00 | 0.16 | 0.08 | 0.002 |
| Isosphaeraceae | 0.00 | 0.00 | 0.13 | 0.06 | 0.001 |
| Alcaligenaceae | 0.01 | 0.00 | 0.12 | 0.04 | 0.007 |
| Nocardiaceae | 0.01 | 0.01 | 0.11 | 0.04 | 0.006 |
| Brucellaceae | 0.00 | 0.00 | 0.11 | 0.03 | 0.001 |
| Aurantimonadaceae | 0.00 | 0.00 | 0.10 | 0.04 | 0.001 |